Precision targeting of β-catenin induces tumor reprogramming and immunity in hepatocellular cancers
- PMID: 40442146
- PMCID: PMC12122713
- DOI: 10.1038/s41467-025-60457-2
Precision targeting of β-catenin induces tumor reprogramming and immunity in hepatocellular cancers
Abstract
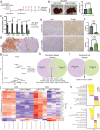
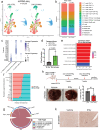
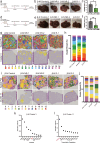
First-line immune checkpoint inhibitor (ICI) combinations show responses in subsets of hepatocellular carcinoma (HCC) patients. Nearly half of HCCs are Wnt-active with mutations in CTNNB1 (encoding for β-catenin), AXIN1/2, or APC, and demonstrate heterogeneous and limited benefit to ICI due to an immune excluded tumor microenvironment. We show significant tumor responses in multiple β-catenin-mutated immunocompetent HCC models to a novel siRNA encapsulated in lipid nanoparticle targeting CTNNB1 (LNP-CTNNB1). Both single-cell and spatial transcriptomics reveal cellular and zonal reprogramming, along with activation of immune regulatory transcription factors IRF2 and POU2F1, re-engaged type I/II interferon signaling, and alterations in both innate and adaptive immunity upon β-catenin suppression with LNP-CTNNB1 at early- and advanced-stage disease. Moreover, ICI enhances response to LNP-CTNNB1 in advanced-stage disease by preventing T cell exhaustion and through formation of lymphoid aggregates (LA). In fact, expression of an LA-like gene signature prognosticates survival for patients receiving atezolizumab plus bevacizumab in the IMbrave150 phase III trial and inversely correlates with CTNNB1-mutatational status in this patient cohort. In conclusion, LNP-CTNNB1 is efficacious as monotherapy and in combination with ICI in CTNNB1-mutated HCCs through impacting tumor cell-intrinsic signaling and remodeling global immune surveillance, providing rationale for clinical investigations.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: S.P.M. has received research grants from Alnylam Pharmaceuticals. He also received funding from Fog Pharmaceuticals and is a consultant on Advisory Boards for Surrozen, AntlerA, Alnylam, Mermaid Bio, Vicero Inc., and UbiquiTx, and there is no pertinent conflict of interest of these entities as relevant to the current manuscript. T.D., M.M., and W.B. are employed by Alnylam Pharmaceuticals, Cambridge, MA. X.G., H.K., and Y.G. are employed by Genentech Inc., San Francisco, CA. No other authors have any relevant conflicts of interests to declare regarding the current study.
Figures





Update of
-
Precision targeting of β-catenin induces tumor reprogramming and immunity in hepatocellular cancers.Res Sq [Preprint]. 2024 Dec 12:rs.3.rs-5494074. doi: 10.21203/rs.3.rs-5494074/v1. Res Sq. 2024. Update in: Nat Commun. 2025 May 30;16(1):5009. doi: 10.1038/s41467-025-60457-2. PMID: 39711542 Free PMC article. Updated. Preprint.
References
-
- Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin.71, 209–249 (2021). - PubMed
-
- Cheng, A. L. et al. Updated efficacy and safety data from IMbrave150: Atezolizumab plus bevacizumab vs. sorafenib for unresectable hepatocellular carcinoma. J. Hepatol.76, 862–873 (2022). - PubMed
-
- Sangro, B. et al. Four-year overall survival update from the phase III HIMALAYA study of tremelimumab plus durvalumab in unresectable hepatocellular carcinoma. Ann. Oncol. 35, 448–457 (2024). - PubMed
-
- Rimassa, L., Finn, R. S. & Sangro, B. Combination immunotherapy for hepatocellular carcinoma. J. Hepatol.79, 506–515 (2023). - PubMed
MeSH terms
Substances
Grants and funding
- F30 CA284540/CA/NCI NIH HHS/United States
- UM1 CA186690/CA/NCI NIH HHS/United States
- R01 DK062277/DK/NIDDK NIH HHS/United States
- P30 DK120531/DK/NIDDK NIH HHS/United States
- R01 CA250227/CA/NCI NIH HHS/United States
- P30CA047904/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- F30CA284540/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R01CA251155/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- UM1CA186690/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- P30 CA047904/CA/NCI NIH HHS/United States
- R01CA250227/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R01 CA251155/CA/NCI NIH HHS/United States
- T32 EB001026/EB/NIBIB NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

