Integrated multi-omics reveals different host crosstalk of atopic dermatitis-enriched Bifidobacterium longum Strains
- PMID: 40442154
- PMCID: PMC12122682
- DOI: 10.1038/s41522-025-00714-w
Integrated multi-omics reveals different host crosstalk of atopic dermatitis-enriched Bifidobacterium longum Strains
Abstract
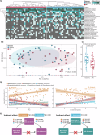
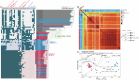
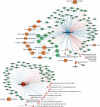
The infant gut microbiome is essential for long-term health and is linked to atopic dermatitis (AD), although the underlying mechanisms are not fully understood. This study investigated gut microbiome-host interactions in 31 infants with AD and 29 healthy controls using multi-omics approaches, including metagenomic, host transcriptomic, and metabolomic analyses. Microbial diversity was significantly altered in AD, with Bifidobacterium longum and Clostridium innocuum associated with these changes. At the strain-level, only B. longum differed significantly between groups, with pangenome analyses identifying genetic variations potentially affecting amino acid and lipid metabolites. Notably, B. longum subclade I, which was more prevalent in healthy controls, correlated with host transcriptomic pathways involved in phosphatidylinositol 3-kinase-AKT signaling and neuroactive ligand-receptor pathways, as well as specific metabolites, including tetrahydrocortisol and ornithine. These findings highlight the role of B. longum strain-level variation in infants, offering new insights into microbiome-host interactions related to AD.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
References
-
- Langan S., Irvine A., Weidinger S. Atopic dermatitisLANCET. 396, 758–758 (2020). - PubMed
-
- Huang, Y. J. et al. The microbiome in allergic disease: Current understanding and future opportunities-2017 PRACTALL document of the American Academy of Allergy, Asthma & Immunology and the European Academy of Allergy and Clinical Immunology. J. Allergy Clin. Immunol.139, 1099–1110 (2017). - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

