Cryo-EM structures of HCV E2 glycoprotein bound to neutralizing and non-neutralizing antibodies determined using bivalent Fabs as fiducial markers
- PMID: 40442315
- PMCID: PMC12122859
- DOI: 10.1038/s42003-025-08239-w
Cryo-EM structures of HCV E2 glycoprotein bound to neutralizing and non-neutralizing antibodies determined using bivalent Fabs as fiducial markers
Abstract
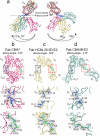
Global elimination of hepatitis C virus (HCV) will require an effective cross-genotype vaccine. The HCV E2 envelope glycoprotein is the main target of neutralizing antibodies but also contains epitopes that elicit non-neutralizing antibodies which may provide protection through Fc effector functions rather than direct neutralization. We determined cryo-EM structures of a broadly neutralizing antibody, a moderately neutralizing antibody, and a non-neutralizing antibody bound to E2 to resolutions of 3.8, 3.3, and 3.7 Å, respectively. Whereas the broadly neutralizing antibody targeted the front layer of E2 and the non-neutralizing antibody targeted the back layer, the moderately neutralizing antibody straddled both front and back layers, and thereby defined a new neutralizing epitope on E2. The small size of complexes between conventional (monovalent) Fabs and E2 (~110 kDa) presented a challenge for cryo-EM. Accordingly, we engineered bivalent versions of E2-specific Fabs that doubled the size of Fab-E2 complexes and conferred highly identifiable shapes to the complexes that facilitated particle selection and orientation for image processing. This study validates bivalent Fabs as new fiducial markers for cryo-EM analysis of small proteins such as HCV E2 and identifies a new target epitope for vaccine development.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






References
-
- Global Hepatitis Reporthttps://www.globalhep.org/sites/default/files/content/resources/files/20... (2024).
-
- Rosen, H. R. “HepC, where art thou?”: What are the remaining (fundable) questions in hepatitis C virus research? Hepatology65, 341–349 (2017). - PubMed
-
- Micallef, J. M., Kaldor, J. M. & Dore, G. J. Spontaneous viral clearance following acute hepatitis C infection: a systematic review of longitudinal studies. J. Viral Hepat.13, 34–41 (2006). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

