Distinct tau filament folds in human MAPT mutants P301L and P301T
- PMID: 40442318
- PMCID: PMC12350173
- DOI: 10.1038/s41594-025-01575-9
Distinct tau filament folds in human MAPT mutants P301L and P301T
Abstract
Mutations in MAPT, the tau gene, give rise to frontotemporal dementia and parkinsonism linked to chromosome 17 (FTDP-17), with abundant filamentous tau inclusions in brain cells. Mutations that encode missense variants of residue P301 are the most common and result in the formation of filamentous inclusions made of mutant four-repeat tau. Here we report the cryo-electron microscopy structures of tau filaments from five individuals belonging to three different families with mutation P301L and from one individual from a family with mutation P301T. A distinct three-lobed tau fold resembling the two-layered fold of Pick's disease was present in the individuals with P301L tau. Two different tau folds were found in the individual with mutation P301T, the less abundant of which was a variant of the three-lobed fold. The major P301T tau fold was V-shaped, with partial similarity to the four-layered tau folds of corticobasal degeneration and argyrophilic grain disease.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


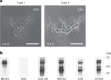



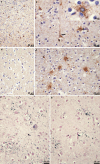


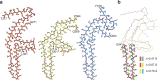
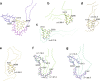


Update of
-
Novel tau filament folds in individuals with MAPT mutations P301L and P301T.bioRxiv [Preprint]. 2024 Aug 17:2024.08.15.608062. doi: 10.1101/2024.08.15.608062. bioRxiv. 2024. Update in: Nat Struct Mol Biol. 2025 Aug;32(8):1470-1478. doi: 10.1038/s41594-025-01575-9. PMID: 39185206 Free PMC article. Updated. Preprint.
References
-
- Goedert, M., Spillantini, M. G., Jakes, R., Rutherford, D. & Crowther, R. A. Multiple isoforms of human microtubule-associated protein tau: sequences and localization in neurofibrillary tangles of Alzheimer’s disease. Neuron3, 519–524 (1989). - PubMed
-
- Scheres, S. H. W., Ryskeldi-Falcon, B. & Goedert, M. Molecular pathology of neurodegenerative diseases by cryo-EM of amyloids. Nature621, 701–710 (2023). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

