The transcriptomes, connections and development of submucosal neuron classes in the mouse small intestine
- PMID: 40442499
- PMCID: PMC12148937
- DOI: 10.1038/s41593-025-01962-x
The transcriptomes, connections and development of submucosal neuron classes in the mouse small intestine
Abstract
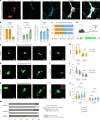
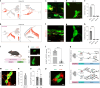
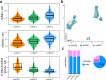
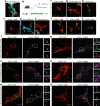
The enteric submucosal plexus regulates essential digestive functions, yet its neuronal composition remains incompletely understood. We identified two putative secretomotor neuron classes and a previously unrecognized submucosal intrinsic primary afferent neuron class through single-cell RNA sequencing in the mouse small intestine. Using viral-mediated labeling of each class, we uncovered their morphologies and neural projections in the submucosa-mucosa context, finding connections among all classes and an unexpected close association with enterochromaffin cells. Further transcriptome analysis at the postnatal stage and lineage tracing revealed that neuron identities in the submucosal plexus emerge through an initial binary fate split at neurogenesis, followed by phenotypic diversification, akin to the developmental process of the myenteric plexus. We propose a unified developmental framework for neuronal diversification across the gut wall. Our study offers comprehensive molecular, developmental and morphological insights into submucosal neurons, opening new avenues for exploring physiological functions, circuit dynamics and formation of the submucosal plexus.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures












References
-
- Furness, J. B. The enteric nervous system and neurogastroenterology. Nat. Rev. Gastroenterol. Hepatol.9, 286–294 (2012). - PubMed
-
- Furness, J. B. The Enteric Nervous System, Second Edition (Blackwell Publishing, 2006).
-
- Vanner, S. & Macnaughton, W. K. Submucosal secretomotor and vasodilator reflexes. Neurogastroenterol. Motil.16, 39–43 (2004). - PubMed
-
- Neckel, P. H. Annotated translation of Georg Meissner’s first description of the submucosal plexus. Neurogastroenterol. Motil.35, e14480 (2023). - PubMed
MeSH terms
Grants and funding
- R01 DK129315/DK/NIDDK NIH HHS/United States
- 2020-01129; 2022-01570/Vetenskapsrådet (Swedish Research Council)
- FO2023-0130; FO2021-0105/Hjärnfonden (Swedish Brain Foundation)
- divENSify 101045026/EC | EC Seventh Framework Programm | FP7 Ideas: European Research Council (FP7-IDEAS-ERC - Specific Programme: "Ideas" Implementing the Seventh Framework Programme of the European Community for Research, Technological Development and Demonstration Activities (2007 to 2013))
- 2020.0109/Knut och Alice Wallenbergs Stiftelse (Knut and Alice Wallenberg Foundation)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

