Targeting BATF2-RGS2 axis reduces T-cell exhaustion and restores anti-tumor immunity
- PMID: 40442751
- PMCID: PMC12123873
- DOI: 10.1186/s12943-025-02351-5
Targeting BATF2-RGS2 axis reduces T-cell exhaustion and restores anti-tumor immunity
Erratum in
-
Correction: Targeting BATF2-RGS2 axis reduces T-cell exhaustion and restores anti-tumor immunity.Mol Cancer. 2025 Jun 23;24(1):182. doi: 10.1186/s12943-025-02387-7. Mol Cancer. 2025. PMID: 40551135 Free PMC article. No abstract available.
Abstract
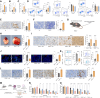
Objective: This study aims to investigate the role of RGS2 in immune regulation in lung cancer (LC) and explore the regulatory relationship between RGS2 and BATF2 in modulating T cell exhaustion and tumor immune evasion.
Methods: Single-cell transcriptome-based analysis was performed to identify CD8+ T-cell profiles and regulatory factors in six LC patients receiving neoadjuvant PD-1 blockade therapy. Mouse 3LL cells or murine tumor organoid models were transplanted into wild-type, RGS2 knock-out (RGS2-/-), or BATF2 knock-out (BATF2-/-) mice to analyze the effects of RGS2 and BATF2 on tumor growth, metastasis, and immune cell infiltration. CD8+ from these mice were isolated and co-cultured with cancer cells to analyze T cell cytotoxicity in vitro. The transcriptional regulation of RGS2 by BATF2 was analyzed using luciferase reporter assays.
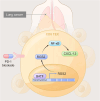
Results: RGS2 was highly expressed in CD8+ T-exhausted (Tex) cells and was associated with pro-inflammatory pathways. High RGS2 expression predicted poor clinical outcomes and limited response to PD-1/PD-L1 blockade therapy. In RGS2-/- mice, tumor metastasis and angiogenesis were suppressed, CD8+ effector T cells were enhanced, and T cell exhaustion markers were reduced. BATF2 was identified as a key transcriptional regulator of RGS2, promoting T cell exhaustion through inhibition of CXCL13 secretion. Knockdown of BATF2 or RGS2 impaired lung cancer cell proliferation and enhanced sensitivity to NK cell-mediated cytotoxicity in vitro. In BATF2-/- mice, the populations of immune active CD8+ T cells were increased, while exhausted T cells were reduced, leading to improved anti-tumor immune responses.
Conclusions: RGS2, regulated by BATF2, plays a critical role in driving T cell exhaustion and tumor immune evasion in LC. Targeting the BATF2-RGS2 axis may enhance the effectiveness of immunotherapy by reversing T cell exhaustion and improving anti-tumor immunity.
Keywords: BATF; CXCL13; Immunosuppression; Lung cancer; RGS2; T-cell exhaustion.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study and included experimental procedures were approved by the institutional animal care and use committee of Zhongda Hospital, School of Medicine, Southeast University (approval NO.20210301085). All animal housing and experiments were conducted in strict accordance with the institutional guidelines for the care and use of laboratory animals. Competing interests: The authors declare no competing interests.
Figures







References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

