Protective effect of phosphoenolpyruvate carboxykinase 1 on inflammation and fibrotic progression of IgA nephropathy
- PMID: 40442892
- PMCID: PMC12128133
- DOI: 10.1080/0886022X.2025.2508297
Protective effect of phosphoenolpyruvate carboxykinase 1 on inflammation and fibrotic progression of IgA nephropathy
Abstract
Introduction: Phosphoenolpyruvate carboxykinase 1 (PCK1) is an essential enzyme of the gluconeogenic pathway, which can affect kidney physiology in various ways. Nevertheless, its role in the progression of IgA nephropathy (IgAN) remains to be elucidated.
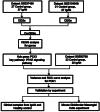
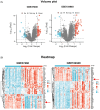
Methods: We identified the differentially expressed genes in the glomeruli of IgAN patients through weighted gene co-expression network analysis across three datasets. Through clinical renal pathological tissue and cellular experiments, we further validated the gene and investigated its relationship with the inflammatory markers and fibrosis indicators in IgAN.
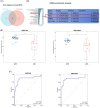
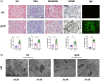
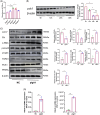
Results: Compared to peritumoral normal tissues, the peroxisome proliferator-activated receptor γ (PPARγ) signaling pathway have been identified as key elements in IgAN pathogenesis from two GEO databases and a validation dataset. PCK1 was identified and validated as one of the most promising candidate genes. The expression of PCK1 in clinically collected kidney specimens was significantly downregulated in patients with IgAN compared to healthy controls. The expression of PCK1 was inversely correlated with clinical indicators such as urinary albumin-to-creatinine ratio, 24-hour proteinuria. In experiments with SV40-transformed mouse glomerular mesangial cells (MCs), PCK1 and PPARγ protein expression levels were significantly decreased in polymeric IgA1 (pIgA1)-stimulated MCs, which contrasts with the increased expression of inflammatory and fibrotic factors. Overexpression of PCK1 inhibited cellular inflammation and fibrotic changes induced by pIgA1, demonstrating protective effects against cellular fibrosis similar to rosiglitazone.
Conclusion: PCK1 exerted a pronounced inhibitory effect on mesangial cell inflammatory markers and fibrosis indicators in IgAN, potentially offering a novel therapeutic target for its treatment.
Keywords: IgA nephropathy; PCK1; PPARγ; fibrosis; inflammation.
Conflict of interest statement
No potential conflicts of interest was reported by the authors.
Figures






Similar articles
-
Downregulation of CD36 alleviates IgA nephropathy by promoting autophagy and inhibiting extracellular matrix accumulation in mesangial cells.Int Immunopharmacol. 2025 Jan 10;144:113672. doi: 10.1016/j.intimp.2024.113672. Epub 2024 Nov 30. Int Immunopharmacol. 2025. PMID: 39616852
-
Regulatory role of the mTOR signaling pathway in autophagy and mesangial proliferation in IgA nephropathy.Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2024 Aug 28;49(8):1220-1231. doi: 10.11817/j.issn.1672-7347.2024.240209. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2024. PMID: 39788511 Free PMC article.
-
RNA-Seq profiling of microdissected glomeruli identifies potential biomarkers for human IgA nephropathy.Am J Physiol Renal Physiol. 2020 Nov 1;319(5):F809-F821. doi: 10.1152/ajprenal.00037.2020. Epub 2020 Sep 21. Am J Physiol Renal Physiol. 2020. PMID: 32954852
-
Transferrin receptor engagement by polymeric IgA1 induces receptor expression and mesangial cell proliferation: role in IgA nephropathy.Contrib Nephrol. 2007;157:144-7. doi: 10.1159/000102457. Contrib Nephrol. 2007. PMID: 17495453 Review.
-
Molecular Insights into the Pathogenesis of IgA Nephropathy.Trends Mol Med. 2015 Dec;21(12):762-775. doi: 10.1016/j.molmed.2015.10.003. Epub 2015 Nov 21. Trends Mol Med. 2015. PMID: 26614735 Review.
References
-
- Barbour SJ, Cattran DC, Kim SJ, et al. . Individuals of Pacific Asian origin with IgA nephropathy have an increased risk of progression to end-stage renal disease. Kidney Int. 2013;84(5):1017–1024. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
