Human microbiota influence the immune cell composition and gene expression in the tumor environment of a murine model of glioma
- PMID: 40443227
- PMCID: PMC12128662
- DOI: 10.1080/19490976.2025.2508432
Human microbiota influence the immune cell composition and gene expression in the tumor environment of a murine model of glioma
Abstract
Background: Immunotherapy has shown success against other cancers but not glioblastoma. Previous data has revealed that microbiota influences anti-PD-1 efficacy. We have previously found that, when using gnotobiotic mice transplanted with human fecal microbiota, the gut microbial composition influenced the response to anti-PD-1 in a mouse model of glioma. However, the role of the human microbiota in influencing the mouse immune cells in the glioma microenvironment and anti-PD-1 response was largely unknown. Using two distinct humanized microbiome (HuM) lines, we used single-cell RNA sequencing (scRNA-seq) to determine how gut microbiota affect immune infiltration and gene expression in a murine glioma model.
Methods: 16S rRNA sequencing was performed on fecal samples from HuM1 (H1) and HuM2 (H2) mice. Mice were intracranially injected with murine glioma cells (GL261), and on day 13 treated with one dose of isotype control or anti-PD1. Mice were euthanized on day 14 for analysis of all immune cells in the tumors by scRNA-seq.
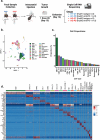
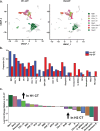
Results: HuM1 and HuM2 mice had different microbial populations, with HuM1 being primarily dominated via Alistipes, and HuM2 being primarily composed of Odoribacter. Sc-RNA-seq of the tumor immune cells revealed 21 clusters with significant differences between H1 and H2 samples with a larger population of M1 type macrophages in H1 samples. Gene expression analysis revealed higher expression of inflammatory markers in the M1 population in H2 mice treated with anti-PD-1.
Conclusions: Microbial gut communities influence the presence and gene activation patterns of immune cells in the brain tumors of mice both under control (isotype) and following anti-PD-1 treatment.
Keywords: Microbiome; glioblastoma; immunotherapy; macrophage; scRNA-seq.
Plain language summary
Microbiota differences influenced numbers and activation of immune cells in a murine glioma model.One day after anti-PD-1 treatment, infiltrating inflammatory macrophages (MM1) increased significantly.HuM2 expressed larger amounts of Nos2 in MM1 cells.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures




References
-
- Lim M, Weller M, Idbaih A, Steinbach J, Finocchiaro G, Raval RR, Ansstas G, Baehring J, Taylor JW, Honnorat J, et al. Phase III trial of chemoradiotherapy with temozolomide plus nivolumab or placebo for newly diagnosed glioblastoma with methylated MGMT promoter. Neuro Oncol. 2022;24(11):1935–1949. doi: 10.1093/neuonc/noac116. - DOI - PMC - PubMed
-
- Omuro A, Brandes AA, Carpentier AF, Idbaih A, Reardon DA, Cloughesy T, Sumrall A, Baehring J, van den Bent M, Bähr O, et al. Radiotherapy combined with nivolumab or temozolomide for newly diagnosed glioblastoma with unmethylated MGMT promoter: an international randomized phase III trial. Neuro Oncol. 2023;25(1):123–134. doi: 10.1093/neuonc/noac099. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
