Epigenetic roles of chromatin remodeling complexes in bone biology and the pathogenesis of bone‑related disease (Review)
- PMID: 40444490
- PMCID: PMC12140095
- DOI: 10.3892/ijmm.2025.5556
Epigenetic roles of chromatin remodeling complexes in bone biology and the pathogenesis of bone‑related disease (Review)
Abstract
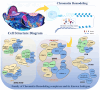
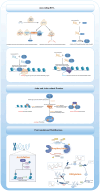
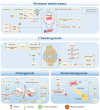
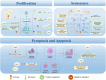
Chromatin remodeling complexes are essential regulators of chromatin architecture, facilitating critical processes such as nucleosome sliding, eviction, histone exchange and post‑translational modifications. By providing an additional layer of epigenetic regulation beyond the canonical genetic code, these complexes significantly influence bone biology and health. Epigenetic regulation through chromatin remodeling complexes is crucial in modulating gene expression and cellular behavior in bone cells. However, alterations in the activity of chromatin remodeling complexes can also contribute to the progression of various bone diseases. Emerging evidence suggests that specific chromatin remodeling factors may serve as potential biomarkers for diagnosing bone‑related conditions and as therapeutic targets for intervention. The present review aims to elucidate the intricate relationship between chromatin remodeling complexes and bone‑related diseases, including osteoporosis, osteoarthritis and osteosarcoma. The present review discusses the diverse subunits of these complexes and their multifaceted roles in regulating key cellular processes such as stemness, differentiation, proliferation, senescence and apoptosis in bone cells. Notably, the present review provides a comprehensive overview of the roles of various chromatin remodeling subunits, such as BRG1, BAF47 and chromodomain‑helicase‑DNA binding 7 (CHD7), in bone metabolism, highlighting their disease‑specific mechanisms, including bromodomain‑containing protein (BRD)9‑mediated pyroptosis in intervertebral disc degeneration and CHD7‑driven bone‑fat imbalance. Furthermore, the present review highlights the therapeutic potential of targeting dysfunctional subunits (such as BRD7 in osteosarcoma and SS18 in synovial sarcoma) and propose AI‑driven structural biology approaches to design chemical modulators. The understudied impact of aging on chromatin remodeling activity in bone homeostasis is also underscored, advocating for longitudinal studies to address this gap. Finally, the distinct functions of each chromatin remodeling complex and its specific subunits in the context of bone‑related diseases were also explored, providing a comprehensive understanding of their contributions to both normal bone physiology and pathological conditions.
Keywords: bone biology; bone development; bone‑related diseases; chromatin remodeling complexes; epigenetic regulation.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
