The cyclic di-GMP receptor YcgR links the second messenger with the putrescine quorum sensing system in modulation of Dickeya oryzae motility
- PMID: 40444987
- PMCID: PMC12239582
- DOI: 10.1128/mbio.01016-25
The cyclic di-GMP receptor YcgR links the second messenger with the putrescine quorum sensing system in modulation of Dickeya oryzae motility
Abstract
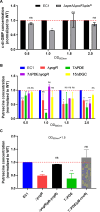
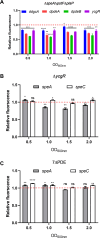
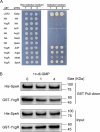
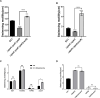
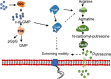
Dickeya oryzae is a prevalent pathogen capable of infecting a variety of crops and plants, and its cell motility plays a key role in invasion of host and subsequent systemic infection. We recently demonstrated that the bacterial second messenger c-di-GMP and the putrescine (PUT)-mediated quorum sensing (QS) system are, respectively, involved in negative and positive regulation of bacterial motility, and vice versa, biofilm formation. In this study, we aimed to investigate the potential interaction of these two signaling mechanisms in the modulation of bacterial motility. The results indicated that null mutation of the PUT system did not seem to have much effect on the cellular level of c-di-GMP; however, deletion of the genes encoding c-di-GMP degradation led to a significant reduction in PUT production. A subsequent study unveiled that the second messenger signaling system interacted with the putrescine QS system through the c-di-GMP receptor YcgR. This interaction enhanced the activity of SpeA, which is the rate-limiting enzyme in the putrescine biosynthesis pathway, resulting in increased intracellular putrescine level. Critically, this facilitative effect was inhibited by c-di-GMP molecules; thus, SpeA, YcgR, and c-di-GMP constitute a regulatory loop modulating D. oryzae motility by controlling the rate of putrescine biosynthesis. The findings from this study provide the first insight into the interaction mechanism between c-di-GMP and putrescine signaling systems in bacteria.IMPORTANCEDickeya oryzae is an important bacterial pathogen that can infect numerous plants and crops, leading to substantial economic losses, especially in rice and banana cultivation. Bacterial motility is a crucial pathogenic factor for D. oryzae as it enables the pathogen to compete for food resources and invade host plants. This motility is negatively regulated by the second messenger c-di-GMP and positively regulated by the quorum sensing signal putrescine (PUT). However, the potential connection between c-di-GMP and PUT signaling systems in regulating the motility of D. oryzae has not been understood. Here, we reveal the link and mechanism of the interaction between them, demonstrating that c-di-GMP interacts with the PUT system via its receptor YcgR. The significance of our research lies in providing the first insight into the molecular interaction between c-di-GMP and PUT signaling networks, both of which are widely conserved signaling mechanisms, and sheds light on the complex and sophisticated regulatory mechanisms that govern bacterial motility and virulence.
Keywords: Dickeya oryzae; YcgR; bacterial motility; c-di-GMP; putrescine.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

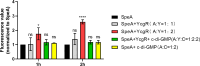
Similar articles
-
Functional analysis of cyclic diguanylate-modulating proteins in Vibrio fischeri.mSystems. 2024 Nov 19;9(11):e0095624. doi: 10.1128/msystems.00956-24. Epub 2024 Oct 22. mSystems. 2024. PMID: 39436151 Free PMC article.
-
Polyethylene terephthalate (PET) primary degradation products affect c-di-GMP-, cAMP-signaling, and quorum sensing (QS) in Vibrio gazogenes DSM 21264.Microbiol Spectr. 2025 Jul;13(7):e0018125. doi: 10.1128/spectrum.00181-25. Epub 2025 Jun 9. Microbiol Spectr. 2025. PMID: 40488468 Free PMC article.
-
The role of cyclic di-GMP in biomaterial-associated infections caused by commensal Escherichia coli.PLoS One. 2025 Aug 20;20(8):e0330229. doi: 10.1371/journal.pone.0330229. eCollection 2025. PLoS One. 2025. PMID: 40833985 Free PMC article.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

