Differential regulation of STING expression and cisplatin sensitivity by autophagy in non-small cell lung cancer cells
- PMID: 40445433
- PMCID: PMC12125072
- DOI: 10.1007/s12032-025-02786-2
Differential regulation of STING expression and cisplatin sensitivity by autophagy in non-small cell lung cancer cells
Abstract
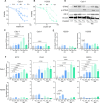
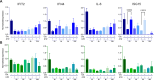
The cGAS-STING pathway is a central signalling mechanism in inflammatory responses and can be activated by cisplatin. Increased autophagic activity has been linked to cisplatin resistance in non-small cell lung cancer (NSCLC); however, how autophagy-STING interactions influence the cisplatin response remains unclear. This study investigates how autophagy modulation affects STING expression and cisplatin sensitivity in NSCLC cells with different basal STING levels. Autophagy was inhibited using chloroquine and induced by serum starvation in Calu-1 and H2030 cells. In Calu-1 cells, cisplatin treatment increased STING expression, activated the cGAS-STING pathway, and induced interferon responses correlated with cisplatin concentration. Autophagy inhibition reduced STING expression and interferon activation while enhancing cisplatin sensitivity. Conversely, autophagy induction caused fluctuations in STING expression and decreased cisplatin sensitivity, with ISG15 expression being selectively increased under serum starvation. In contrast, H2030 cells exhibited low basal STING expression and showed minimal responses to cisplatin or autophagy modulation. These findings suggest that STING expression levels critically influence autophagy-mediated responses to DNA-damaging chemotherapy in NSCLC.
Keywords: Autophagy; Chloroquine; Cisplatin; NSCLC; Starvation; cGAS-STING.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

