Transglutaminase 2 nuclear localization enhances glioblastoma radiation resistance
- PMID: 40445445
- PMCID: PMC12125413
- DOI: 10.1007/s12672-025-02599-9
Transglutaminase 2 nuclear localization enhances glioblastoma radiation resistance
Abstract
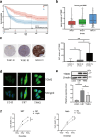
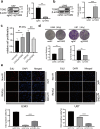
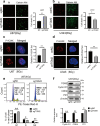
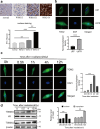
Radiotherapy remains the cornerstone of treatment for glioblastoma (GBM). However, the frequent occurrence of radiation resistance presents a significant therapeutic challenge. A comprehensive understanding of the mechanisms underlying this resistance is essential for improving GBM treatment strategies. In the present study, live-dead cell staining and immunofluorescence staining were employed, and irradiation-resistant cell lines were established. It was observed that transglutaminase 2 (TGM2) plays a pivotal role in enhancing radiation resistance in GBM, facilitating cell proliferation, and promoting DNA damage repair following irradiation. Moreover, immunofluorescence and nucleoplasmic protein extraction assays revealed that TGM2 in GBM rapidly translocates into the nucleus upon irradiation. Through co-immunoprecipitation assays, TGM2 was identified as binding to an increased amount of p53 proteins, thereby promoting p53 degradation post-irradiation. Notably, inhibition of this interaction resulted in a reduction of radiation resistance in GBM. In summary, this study underscores the significance of TGM2 nuclear translocation in radiation resistance and suggests that disrupting TGM2 binding to p53 may offer novel therapeutic insights for overcoming radiation resistance in GBM.
Keywords: DNA damage repair; Glioblastoma; Radiation resistance; Transglutaminase 2; Translocation; p53.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures


References
-
- Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, Hau P, Brandes AA, Gijtenbeek J, Marosi C, Vecht CJ, Mokhtari K, Wesseling P, Villa S, Eisenhauer E, Gorlia T, Weller M, Lacombe D, Cairncross JG, Mirimanoff RO. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–66. 10.1016/S1470-2045(09)70025-7. - DOI - PubMed
-
- Yard BD, Adams DJ, Chie EK, Tamayo P, Battaglia JS, Gopal P, Rogacki K, Pearson BE, Phillips J, Raymond DP, Pennell NA, Almeida F, Cheah JH, Clemons PA, Shamji A, Peacock CD, Schreiber SL, Hammerman PS, Abazeed ME. A genetic basis for the variation in the vulnerability of cancer to DNA damage. Nat Commun. 2016;7:11428. 10.1038/ncomms11428. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
