Environmental DNA adsorption to chitin can promote horizontal gene transfer by natural transformation
- PMID: 40445756
- PMCID: PMC12146716
- DOI: 10.1073/pnas.2420708122
Environmental DNA adsorption to chitin can promote horizontal gene transfer by natural transformation
Abstract
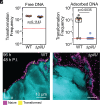
Horizontal gene transfer by natural transformation (NT) is induced in Vibrio cholerae upon attachment to chitin surfaces in the aquatic environment. Here, we show that free environmental DNA adsorbs to chitin surfaces under physiologically realistic conditions. Using live-cell imaging and a fluorescent NT reporter, we demonstrate with cellular resolution microscopy that V. cholerae utilizes chitin-bound DNA as a reservoir for genetic exchange. Additionally, we demonstrate that uptake of DNA from this chitin-bound reservoir requires the forceful retraction of competence type IV pili. These findings uncover a role for retraction force in driving pilus-dependent NT and suggest that chitin particle surfaces can act as hotspots for horizontal gene transfer.
Keywords: Vibrio; biofilm; chitin; natural transformation; pilus retraction force.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures

References
-
- Shapiro B. J., Levade I., Kovacikova G., Taylor R. K., Almagro-Moreno S., Origins of pandemic Vibrio cholerae from environmental gene pools. Nat. Microbiol. 2, 16240 (2016). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

