Application of a metabolic network-based graph neural network for the identification of toxicant-induced perturbations
- PMID: 40445920
- PMCID: PMC12198668
- DOI: 10.1093/toxsci/kfaf065
Application of a metabolic network-based graph neural network for the identification of toxicant-induced perturbations
Abstract
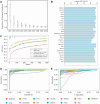
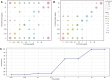
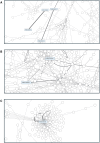
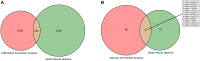
Transcriptomic analyses have been an effective approach to investigate the biological responses and metabolic perturbations by environmental contaminants in rodent models. However, it is well recognized that metabolic networks are highly connected and complex, and that traditional gene expression analysis methods, including pathway analyses, have a limited ability to capture these complexities. Given that metabolism can be effectively represented as a graph, this study aims to apply a network-based graph neural network (GNN) to uncover novel or hidden metabolic perturbations in response to a toxicant. A GNN model based on the mouse Reactome pathways was trained and validated on 7,689 transcriptomic samples from 26 mouse tissues curated from Recount3. This model was then used to identify important reactions in publicly available data from livers of mice treated with the environmental contaminant 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) achieving a performance of 100% when comparing a single dose to a control group. Integrated gradients and centrality analyses identified perturbation of the SUMOylation, cell cycle, P53 signaling, and collagen biosynthesis pathways by TCDD which were not identified using a pathway analysis approach. Collectively, our results demonstrate that GNNs can reveal novel mechanistic insights into toxicant-mediated metabolic disruption, presenting a putative strategy to characterize biological responses to toxicant exposures. Our studies illustrate how the use of a reaction-based graph neural network can support the discovery of toxicant-induced metabolic perturbations, and highlight strengths and challenges in the application of artificial intelligence methods for environmental health research.
Keywords: artificial intelligence; graph neural networks; metabolism; mice; toxicogenetics.
© The Author(s) 2025. Published by Oxford University Press on behalf of the Society of Toxicology.
Figures
Similar articles
-
Assessing the comparative effects of interventions in COPD: a tutorial on network meta-analysis for clinicians.Respir Res. 2024 Dec 21;25(1):438. doi: 10.1186/s12931-024-03056-x. Respir Res. 2024. PMID: 39709425 Free PMC article. Review.
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
-
Accelerated prediction of molecular properties for per- and polyfluoroalkyl substances using graph neural networks with adjacency-free message passing.Environ Pollut. 2025 Oct 1;382:126705. doi: 10.1016/j.envpol.2025.126705. Epub 2025 Jun 30. Environ Pollut. 2025. PMID: 40602636
-
Immunogenicity and seroefficacy of pneumococcal conjugate vaccines: a systematic review and network meta-analysis.Health Technol Assess. 2024 Jul;28(34):1-109. doi: 10.3310/YWHA3079. Health Technol Assess. 2024. PMID: 39046101 Free PMC article.
-
The use of Open Dialogue in Trauma Informed Care services for mental health consumers and their family networks: A scoping review.J Psychiatr Ment Health Nurs. 2024 Aug;31(4):681-698. doi: 10.1111/jpm.13023. Epub 2024 Jan 17. J Psychiatr Ment Health Nurs. 2024. PMID: 38230967
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

