Integrative transcriptomic analysis reveals alternative splicing complexity and transcriptomic diversity in porcine placentas across altitudes
- PMID: 40445954
- PMCID: PMC12137898
- DOI: 10.1093/dnares/dsaf008
Integrative transcriptomic analysis reveals alternative splicing complexity and transcriptomic diversity in porcine placentas across altitudes
Abstract
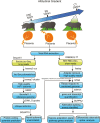
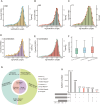
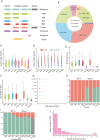
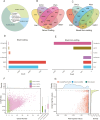
High-altitude hypoxia provides a natural laboratory for studying adaptation in plateau mammals. As an interface for oxygen and nutrient exchange, placenta plays a critical role in fetal development. While high-altitude adaptation in systemic physiological responses and cardiopulmonary tissues has been well-studied, a comprehensive landscape of porcine placental transcriptomic diversity and alternative splicing (AS) complexity across altitudes remains lacking. Here, we integrated Iso-Seq and RNA-Seq to profile placental transcriptomes from placentas of 3 pig breeds across altitudes: the Diannan small-ear pig (DSE ~500 m), the Baoshan pig (BS ~1500 m), and the Changdu Tibetan Pig (CT ~3500 m). We identified 39,776 full-length transcripts, including 25,471 novel ones, significantly enhancing pig genome annotation. Additionally, 24,879 AS events from 8,390 AS genes were detected, with skipping exon (SE) as the most prevalent AS type. Differential expression (DE) and differential alternative splicing (DAS) analyses highlighted key DEGs (IGF1, GHR, RASGRP4, MECOM, SPP1), as well as DAS genes (HIF1A, HSPA8, RHOA, HMGCR, PLAGL1), which may be implicated in placental adaptation to high-altitude conditions. This study provides a comprehensive analysis of the transcriptomic diversity and AS complexity in porcine placentas across altitudes, laying a foundation for future investigations into the molecular mechanisms underlying high-altitude adaptation in plateau mammals.
Keywords: alternative splicing (AS); high-altitude adaptation; pig; placenta; transcriptomics.
© The Author(s) 2025. Published by Oxford University Press on behalf of Kazusa DNA Research Institute.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Maltepe E, Fisher SJ.. Placenta: the forgotten organ. Annu Rev Cell Dev Biol. 2015:31:523–552. https://doi.org/10.1146/annurev-cellbio-100814-125620 - DOI - PubMed
-
- Burton GJ, Fowden AL.. The placenta: a multifaceted, transient organ. Philos Trans R Soc London Ser B. 2015:370:20140066. https://doi.org/10.1098/rstb.2014.0066 - DOI - PMC - PubMed
-
- Gude NM, Roberts CT, Kalionis B, King RG.. Growth and function of the normal human placenta. Thromb Res. 2004:114:397–407. https://doi.org/10.1016/j.thromres.2004.06.038 - DOI - PubMed
-
- Cross JC, Werb Z, Fisher SJ.. Implantation and the placenta: key pieces of the development puzzle. Science. 1994:266:1508–1518. https://doi.org/10.1126/science.7985020 - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

