Probing orthobunyavirus reassortment using Bunyamwera and Batai viruses as models
- PMID: 40445997
- PMCID: PMC12169594
- DOI: 10.1371/journal.pntd.0013120
Probing orthobunyavirus reassortment using Bunyamwera and Batai viruses as models
Abstract
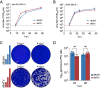
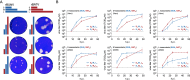
Reassortment is a critical evolutionary mechanism for segmented viruses, enabling the exchange of intact genome segments during co-infection and driving orthobunyavirus evolution; however, the molecular mechanisms underpinning this process remain unclear. With over 100 orthobunyavirus species, many of which are significant human and veterinary pathogens, understanding how reassortment influences transmissibility and virulence is essential for preempting the emergence of novel pathogens. Here, we use Bunyamwera virus (BUNV) and Batai virus (BATV) as models to explore orthobunyavirus reassortment through reverse genetics. We established the first reverse genetics system for BATV, generated reassortants, and employed minigenome assays to assess replication machinery compatibility. Additionally, we developed a novel hybridization chain reaction assay for high-resolution visualization of viral RNA segments. Our findings revealed that all six reassortants between BUNV and BATV are viable, exhibiting notable phenotypic differences in interferon-deficient (IFNAR-/-) mice. This work introduces essential tools and new insights into orthobunyavirus reassortment and pathogenesis, laying the groundwork for understanding this critical evolutionary process.
Copyright: © 2025 Bowen et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Barr JN, Fearns R. Genetic instability of RNA viruses. Genome stability. 2016. p. 21–35.
-
- Elliott RM. Orthobunyaviruses: recent genetic and structural insights. Nature Publishing Group. 2014;12. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

