Human LY9 governs CD4+ T cell IFN-γ immunity to Mycobacterium tuberculosis
- PMID: 40446017
- PMCID: PMC12242830
- DOI: 10.1126/sciimmunol.ads7377
Human LY9 governs CD4+ T cell IFN-γ immunity to Mycobacterium tuberculosis
Abstract
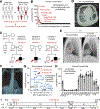
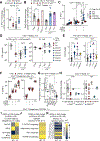
CD4+ T cells are indispensable for optimal immunity to Mycobacterium tuberculosis (M.tb), a pathogen that triggers tuberculosis (TB) in humans. M.tb-specific human CD4+ T cells are known to polarize toward an interferon-γ (IFN-γ)-producing, CCR4-CCR6+CXCR3+T-bet+RORγT+ T helper 1* cell (TH1*cell) memory phenotype. We report that autosomal recessive deficiency of the human lymphocytic surface receptor LY9 (SLAMF3 and CD229), which is found in less than 10-5 individuals in the general population, underlies TB in three unrelated patients due to selective impairment in IFN-γ production by TH1* cells. TH1* cells express higher levels of LY9 than other CD4+ T cells. Mechanistically, LY9 polarizes naïve CD4+ T cells toward memory TH1* cells by inducing T-bet via signaling lymphocytic activation molecule (SLAM)-associated protein (SAP) and RORγT (thymus-specific retinoid-related orphan receptor γ) without SAP. LY9 costimulation enhances TCR-driven IFN-γ production of memory TH1*, but not TH1, cells in a T cell-intrinsic manner via NFAT1 (nuclear factor of activated T cells 1) and RORγT. LY9 is likely to govern an optimal TH1* cell- and IFN-γ-dependent protective immunity to M.tb in humans.
Conflict of interest statement
Competing interests
The authors declare that there are no relevant competing interests.
Figures




References
-
- WHO, Global Tuberculosis Report 2022 (2022).
-
- Vynnycky E, Fine PEM, Lifetime Risks, Incubation Period, and Serial Interval of Tuberculosis. Am. J. Epidemiol. 152, 247–263 (2000). - PubMed
-
- Kallmann FJ, Reisner D, Twin Studies on the Significance of Genetic Factors in Tuberculosis,. Am. Rev. Tuberc. 47, 549–574 (1943).
-
- Comstock GW, Tuberculosis in twins: a re-analysis of the Prophit survey. Am. Rev. Respir. Dis. 117, 621–4 (1978). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

