Covalent Adducts Formed by the Androgen Receptor Transactivation Domain and Small Molecule Drugs Remain Disordered
- PMID: 40446154
- PMCID: PMC12199303
- DOI: 10.1021/acs.jcim.5c00833
Covalent Adducts Formed by the Androgen Receptor Transactivation Domain and Small Molecule Drugs Remain Disordered
Abstract
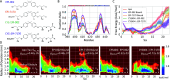
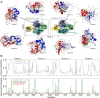
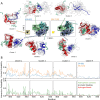
Intrinsically disordered proteins are implicated in many human diseases. Small molecules that target the disordered androgen receptor transactivation domain have entered human trials for the treatment of castration-resistant prostate cancer. These molecules have been shown to react with cysteine residues of the androgen receptor transactivation domain and form covalent adducts under physiological conditions. It is currently unclear how the covalent attachment of these molecules alters the conformational ensemble of the androgen receptor. Here, we utilize all-atom molecular dynamics computer simulations to simulate covalent adducts of small molecule ligands EPI-002 and EPI-7170 bound to the disordered androgen receptor transactivation domain. Our simulations reveal that the conformational ensembles of androgen receptor transactivation domain covalent adducts are heterogeneous and disordered. We find that covalent attachment of EPI-002 and EPI-7170 increases the population of collapsed helical transactivation domain conformations relative to the populations observed in non-covalent binding simulations, and we identify networks of protein-ligand interactions that stabilize collapsed conformations in covalent adduct ensembles. We compare the populations of protein-ligand interactions observed in covalent adduct ensembles to those observed in non-covalent ligand-bound ensembles and find substantial differences. Our results provide atomically detailed descriptions of covalent adducts formed by small molecules and an intrinsically disordered protein and suggest strategies for developing more potent covalent inhibitors of intrinsically disordered proteins.
Figures

Update of
-
Covalent adducts formed by the androgen receptor transactivation domain and small molecule drugs remain disordered.bioRxiv [Preprint]. 2024 Nov 15:2024.11.12.623257. doi: 10.1101/2024.11.12.623257. bioRxiv. 2024. Update in: J Chem Inf Model. 2025 Jun 23;65(12):6221-6237. doi: 10.1021/acs.jcim.5c00833. PMID: 39605539 Free PMC article. Updated. Preprint.
Similar articles
-
Covalent adducts formed by the androgen receptor transactivation domain and small molecule drugs remain disordered.bioRxiv [Preprint]. 2024 Nov 15:2024.11.12.623257. doi: 10.1101/2024.11.12.623257. bioRxiv. 2024. Update in: J Chem Inf Model. 2025 Jun 23;65(12):6221-6237. doi: 10.1021/acs.jcim.5c00833. PMID: 39605539 Free PMC article. Updated. Preprint.
-
Covalent Destabilizing Degrader of AR and AR-V7 in Androgen-Independent Prostate Cancer Cells.J Am Chem Soc. 2025 Jun 18;147(24):20512-20524. doi: 10.1021/jacs.5c02801. Epub 2025 Jun 9. J Am Chem Soc. 2025. PMID: 40490871
-
Covalent Destabilizing Degrader of AR and AR-V7 in Androgen-Independent Prostate Cancer Cells.bioRxiv [Preprint]. 2025 Feb 16:2025.02.12.637117. doi: 10.1101/2025.02.12.637117. bioRxiv. 2025. Update in: J Am Chem Soc. 2025 Jun 18;147(24):20512-20524. doi: 10.1021/jacs.5c02801. PMID: 39990458 Free PMC article. Updated. Preprint.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
Cited by
-
Ensemble Docking for Intrinsically Disordered Proteins.J Chem Inf Model. 2025 Jul 14;65(13):6847-6860. doi: 10.1021/acs.jcim.5c00370. Epub 2025 Jun 18. J Chem Inf Model. 2025. PMID: 40532196 Free PMC article.
-
Ensemble docking for intrinsically disordered proteins.bioRxiv [Preprint]. 2025 Jan 26:2025.01.23.634614. doi: 10.1101/2025.01.23.634614. bioRxiv. 2025. Update in: J Chem Inf Model. 2025 Jul 14;65(13):6847-6860. doi: 10.1021/acs.jcim.5c00370. PMID: 39896485 Free PMC article. Updated. Preprint.
References
-
- Camacho-Zarco A. R., Schnapka V., Guseva S., Abyzov A., Adamski W., Milles S., Jensen M. R., Zidek L., Salvi N., Blackledge M.. NMR provides unique insight into the functional dynamics and interactions of intrinsically disordered proteins. Chem. Rev. 2022;122:9331–9356. doi: 10.1021/acs.chemrev.1c01023. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

