The Slx4-Rad1-Rad10 nuclease differentially regulates deletions and duplications induced by a replication fork barrier
- PMID: 40446207
- PMCID: PMC12151478
- DOI: 10.1371/journal.pgen.1011720
The Slx4-Rad1-Rad10 nuclease differentially regulates deletions and duplications induced by a replication fork barrier
Abstract
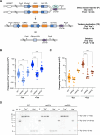
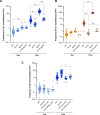
Genome instability is a hallmark of cancer that can be caused by DNA replication stress. Copy number variation (CNV) is a type of genomic instability that has been associated with both tumorigenesis and drug resistance, but how these structural variants form in response to replication stress is not fully understood. Here, we established a direct repeat genetic reporter in Saccharomyces cerevisiae to detect recombination events that result in either a duplication or a deletion. Using this system, we measured recombination resulting from site-specific replication fork stalling initiated by Tus binding to an array of Ter sites. We found that a Tus/Ter fork block downstream of direct repeats induced CNV by a mechanism involving the Mph1 translocase, Exo1-catalyzed end resection and Rad51-dependent strand invasion. While the Slx4 scaffold protein and its nuclease-binding partner, Rad1-Rad10, were shown to be required for duplications, we found that they suppress deletion formation in this context. These opposing functions suggest that both recombination products arise through a large loop heteroduplex intermediate that is cleaved by Rad1-Rad10 in a manner that promotes duplications and eliminates deletions. Taken together, these studies give insight into the mechanisms governing CNV in the context of replication fork stalling, which may ultimately provide a better understanding of how replication stress contributes to cancer and other diseases characterized by genome instability.
Copyright: © 2025 Triplett et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

