Multi-tissue expression and splicing data prioritise anatomical subsite- and sex-specific colorectal cancer susceptibility genes
- PMID: 40447568
- PMCID: PMC12125321
- DOI: 10.1038/s41467-025-60275-6
Multi-tissue expression and splicing data prioritise anatomical subsite- and sex-specific colorectal cancer susceptibility genes
Abstract
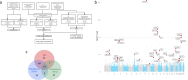
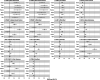
Genome-wide association studies have suggested numerous colorectal cancer (CRC) susceptibility genes, but their causality and therapeutic potential remain unclear. To prioritise causal associations between gene expression/splicing and CRC risk (52,775 cases; 45,940 controls), we perform a transcriptome-wide association study (TWAS) across six tissues with Mendelian randomisation and colocalisation, integrating sex- and anatomical subsite-specific analyses. Here we reveal 37 genes with robust causal links to CRC risk, ten of which have not previously been reported by TWAS. Most likely causal genes with evidence of cancer cell dependency show elevated expression linked to risk, suggesting therapeutic potential. Notably, SEMA4D, encoding a protein targeted by an investigational CRC therapy, emerges as a key risk gene. We also identify a female-specific association with CRC risk for CCM2 expression and subsite-specific associations, including LAMC1 with rectal cancer risk. These findings offer valuable insights into CRC molecular mechanisms and support promising therapeutic avenues.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: T.G.R. is employed full-time by GlaxoSmithKline outside of the research presented in this manuscript. Where authors are identified as personnel of the International Agency for Research on Cancer/World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy, or views of the International Agency for Research on Cancer/World Health Organization. This article is the result of the scientific work of Dr Murphy while he was affiliated at IARC. The remaining authors declare no competing interests.
Figures

References
MeSH terms
Substances
Grants and funding
- MC_UU_00032/03/RCUK | Medical Research Council (MRC)
- C18281/A29019/Cancer Research UK (CRUK)
- P30 CA015704/CA/NCI NIH HHS/United States
- S10 OD028685/OD/NIH HHS/United States
- U01 CA137088/CA/NCI NIH HHS/United States
- U24 CA074794/CA/NCI NIH HHS/United States
- APP1179170/Department of Health | National Health and Medical Research Council (NHMRC)
- U01 CA167551/CA/NCI NIH HHS/United States
- APP177524/Department of Health | National Health and Medical Research Council (NHMRC)
- C18281/A30905/Cancer Research UK (CRUK)
- T32 ES013678/ES/NIEHS NIH HHS/United States
- U01 CA122839/CA/NCI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- P01 CA196569/CA/NCI NIH HHS/United States
- HHSN268201700006C/HL/NHLBI NIH HHS/United States
- U19 CA148107/CA/NCI NIH HHS/United States
- R01 CA059045/CA/NCI NIH HHS/United States
- R01 CA197350/CA/NCI NIH HHS/United States
- R01 CA076366/CA/NCI NIH HHS/United States
- U01 CA074794/CA/NCI NIH HHS/United States
- HHSN268201200008C/HL/NHLBI NIH HHS/United States
- R01 CA242218/CA/NCI NIH HHS/United States
- P30 CA014089/CA/NCI NIH HHS/United States
- 218495/Z/19/Z/Wellcome Trust (Wellcome)
- R01 CA081488/CA/NCI NIH HHS/United States
- R01 CA143237/CA/NCI NIH HHS/United States
- R01 CA201407/CA/NCI NIH HHS/United States
- HHSN268201200008I/HL/NHLBI NIH HHS/United States
- 001/WHO_/World Health Organization/International
LinkOut - more resources
Full Text Sources
Medical

