Circular RNA-mediated inverse prime editing in human cells
- PMID: 40447589
- PMCID: PMC12125189
- DOI: 10.1038/s41467-025-59120-7
Circular RNA-mediated inverse prime editing in human cells
Abstract
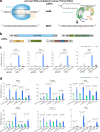
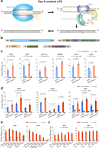
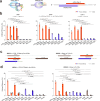
Prime editors are restricted to performing precise edits downstream of cleavage sites, thereby limiting their editing scope. Therefore, we develop inverse prime editors (iPEs) that act upstream of the nickase cleavage site by replacing nCas9-H840A with nCas9-D10A, but the editing efficiencies are limited. To address this limitation, we develop circular RNA-mediated iPEs (ciPEs), achieving editing efficiencies ranging from 0.1% to 24.7%. Further optimization using Rep-X helicase increases editing efficiencies to a range of 2.7%-55.4%. The Rep-X-assisted ciPE system thus expands the scope of editing and improves efficiencies at genomic sites that are previously difficult to target. The Rep-X-assisted ciPE system will complement canonical PE system in enabling more extensive and efficient editing across a wider range of the human genome.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare the following competing interests: C.G. and R.L. have submitted a patent application on the prime editors developed in this work (Application No. 2024112204624). The remaining authors declare no competing interests.
Figures


Similar articles
-
Prime editor with rational design and AI-driven optimization for reverse editing window and enhanced fidelity.Nat Commun. 2025 Jun 3;16(1):5144. doi: 10.1038/s41467-025-60495-w. Nat Commun. 2025. PMID: 40461565 Free PMC article.
-
Enhancing CRISPR prime editing by reducing misfolded pegRNA interactions.Elife. 2024 Jun 7;12:RP90948. doi: 10.7554/eLife.90948. Elife. 2024. PMID: 38847802 Free PMC article.
-
Prime editing using CRISPR-Cas12a and circular RNAs in human cells.Nat Biotechnol. 2024 Dec;42(12):1867-1875. doi: 10.1038/s41587-023-02095-x. Epub 2024 Jan 10. Nat Biotechnol. 2024. PMID: 38200119
-
Latest Developed Strategies to Minimize the Off-Target Effects in CRISPR-Cas-Mediated Genome Editing.Cells. 2020 Jul 2;9(7):1608. doi: 10.3390/cells9071608. Cells. 2020. PMID: 32630835 Free PMC article. Review.
-
Off-Target Editing by CRISPR-Guided DNA Base Editors.Biochemistry. 2019 Sep 10;58(36):3727-3734. doi: 10.1021/acs.biochem.9b00573. Epub 2019 Aug 26. Biochemistry. 2019. PMID: 31433621 Free PMC article. Review.
References
-
- Zhao, Z. et al. Prime editing: advances and therapeutic applications. Trends Biotechnol.41, 1000–1012 (2023). - PubMed
-
- Gao, C. Genome engineering for crop improvement and future agriculture. Cell184, 1621–1635 (2021). - PubMed
-
- Li, B. et al. Targeted genome-modification tools and their advanced applications in crop breeding. Nat. Rev. Genet.25, 603–622 (2024). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

