Real-world performance analysis of a universal computational reasoning model for precision oncology in lung cancer
- PMID: 40447752
- PMCID: PMC12125307
- DOI: 10.1038/s41698-025-00943-4
Real-world performance analysis of a universal computational reasoning model for precision oncology in lung cancer
Abstract
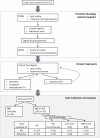
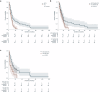
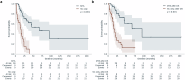
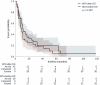
Tumors harbor multiple genetic alterations, yet treatment decisions are commonly based on single biomarkers, leading to underutilization of genomic information by comprehensive molecular tests, uncertainty in clinical practice, and frequent treatment failures. Although molecular tumor boards can assist personalized treatments, this process is not scalable or standardized, resulting in highly discordant recommendations. Validated digital solutions for personalized decision support are highly needed. The Digital Drug Assignment (DDA) system is a computational reasoning model that scores treatment options based on the full tumor genomic data. We retrospectively analyzed data of 111 lung cancer patients and found that high-score MTAs (1000≦DDA score) provided significant clinical benefit over other treatments, in terms of ORR, PFS, and OS. These results demonstrate that the DDA system is predictive of relative benefit of the various agents used in lung cancer care. Digital drug assignment can potentially address challenges with complex molecular profiles in routine clinical settings.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: A.D. employment Genomate Health, stock options Genomate Health, D.L. employment Genomate Health, stock options Genomate Health, M.B. employment Genomate Health, M.K.S. employment Genomate Health, stock options Genomate Health, G.G.K. employment Genomate Health, D.T. employment Genomate Health, stock options Genomate Health, A.T. employment Genomate Health, A.B. employment Oncompass Medicine, R.S.D. employment Genomate Health, stock options Genomate Health, B.V. employment Genomate Health, stock options Genomate Health, E.V. employment Oncompass Medicine, J.D. employment Oncompass Medicine, G.P. employment Oncompass Medicine, D.M. employment Genomate Health, R.S. board member Oncompass Medicine and Genomate Health, has research or advisory contracts with G.E., Janssen (Johnson & Johnson), Danone/Nutricia, Novartis, Sanofi, Biogaia Winclove, ProGastro, and Nestle, M.K. had consulting roles with AZD and Roche, C.R. is an advisory board member at AstraZeneca, Daiichi Sankyo, Regeneron and Novocure, Bristol-Myers Squibb (BMS), Novartis, Invitae, Guardant Health, COR2ED, Bayer, Boehringer Ingelheim, Abbvie, Invitae, Janssen, EMD Serono; has research grants from Astra Zeneca, Thermo Fisher, Oncohost, Lung Cancer Research Foundation, National Foundation for Cancer Research, and U54 (National Institute of Health), has research collaborations with GuardantHealth, Foundation Medicine, Roche Diagnostics, EMD Serono; is a scientific advisory board member of Imagene. C.L.T. participated in advisory boards from MSD, BMS, Merck, Astra Zeneca, Celgene, Seattle Genetics, Roche, Novartis, Rakuten, Nanobiotix, and GSK R.D. employment Genomate Health, stock options Genomate Health, I.P. is an employee and equity holder in Oncompass Medicine Inc., and Genomate Health Inc. D.K., V.K., I.V.N., A.Z.D., L.U. These authors declare no competing interests. Ethical approval: All patients provided written informed consent to use anonymized data for research purposes. Prior to conducting the study, ethics approval was obtained from the National Institute of Pharmacy and Nutrition (approval no. OGYEI/50268/2017), in accordance with the principles of the Declaration of Helsinki.
Figures

References
Grants and funding
- 2019-1.1.1- PIACI-KFI-2019-00367/Nemzeti Kutatási Fejlesztési és Innovációs Hivatal
- 2019-1.1.1- PIACI-KFI-2019-00367/Nemzeti Kutatási Fejlesztési és Innovációs Hivatal
- 2019-1.1.1- PIACI-KFI-2019-00367/Nemzeti Kutatási Fejlesztési és Innovációs Hivatal
- 2019-1.1.1- PIACI-KFI-2019-00367/Nemzeti Kutatási Fejlesztési és Innovációs Hivatal
- 2019-1.1.1- PIACI-KFI-2019-00367/Nemzeti Kutatási Fejlesztési és Innovációs Hivatal
- 2019-1.1.1- PIACI-KFI-2019-00367/Nemzeti Kutatási Fejlesztési és Innovációs Hivatal
- 2019-1.1.1- PIACI-KFI-2019-00367/Nemzeti Kutatási Fejlesztési és Innovációs Hivatal
- 2019-1.1.1- PIACI-KFI-2019-00367/Nemzeti Kutatási Fejlesztési és Innovációs Hivatal
- 2019-1.1.1- PIACI-KFI-2019-00367/Nemzeti Kutatási Fejlesztési és Innovációs Hivatal
- 2019-1.1.1- PIACI-KFI-2019-00367/Nemzeti Kutatási Fejlesztési és Innovációs Hivatal
- 2019-1.1.1- PIACI-KFI-2019-00367/Nemzeti Kutatási Fejlesztési és Innovációs Hivatal
- 2019-1.1.1- PIACI-KFI-2019-00367/Nemzeti Kutatási Fejlesztési és Innovációs Hivatal
- 2019-1.1.1- PIACI-KFI-2019-00367/Nemzeti Kutatási Fejlesztési és Innovációs Hivatal
- 2019-1.1.1- PIACI-KFI-2019-00367/Nemzeti Kutatási Fejlesztési és Innovációs Hivatal
- 2019-1.1.1- PIACI-KFI-2019-00367/Nemzeti Kutatási Fejlesztési és Innovációs Hivatal
- 2019-1.1.1- PIACI-KFI-2019-00367/Nemzeti Kutatási Fejlesztési és Innovációs Hivatal
- 2019-1.1.1- PIACI-KFI-2019-00367/Nemzeti Kutatási Fejlesztési és Innovációs Hivatal
LinkOut - more resources
Full Text Sources
Medical

