The IL-22-oncostatin M axis promotes intestinal inflammation and tumorigenesis
- PMID: 40447860
- PMCID: PMC12133592
- DOI: 10.1038/s41590-025-02149-z
The IL-22-oncostatin M axis promotes intestinal inflammation and tumorigenesis
Abstract
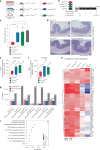
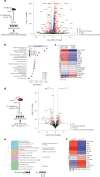
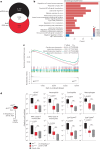
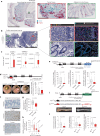
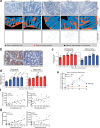
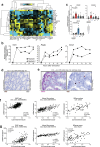
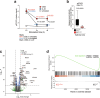
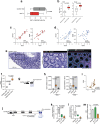
Multicellular cytokine networks drive intestinal inflammation and colitis-associated cancer (CAC). Interleukin-22 (IL-22) exerts both protective and pathogenic effects in the intestine, but the mechanisms that regulate this balance remain unclear. Here, we identify that IL-22 directly induces responsiveness to the IL-6 family cytokine oncostatin M (OSM) in intestinal epithelial cells (IECs) by activating STAT3 and upregulating the OSM receptor. In turn, OSM synergizes with IL-22 to sustain STAT3 activation in IECs and promote proinflammatory epithelial adaptation and immune cell chemotaxis to the inflamed intestine. Conditional deletion of the OSM receptor in IECs protects mice from both colitis and CAC, and pharmacological blockade of OSM attenuates established CAC. Thus, IL-22 and OSM form a pathogenic circuit that drives inflammation and tumorigenesis. Our findings reveal a previously unknown mechanism by which OSM supports intestinal pathology and highlight the IL-22-OSM axis as a promising therapeutic target for inflammatory bowel disease and CAC.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: A.S., B.B., J.E.K., J.H., J.Z., M.K., J.R., L.W., M.E.K. and N.R.W. are current or former members of Genentech. N.R.W. and A.N.H. are inventors of patents relating to OSM as a therapeutic target for IBD (WO 2016/120625 A1). The remaining authors declare no competing interests.
Figures










References
MeSH terms
Substances
Grants and funding
- 92087/Volkswagen Foundation (VolkswagenStiftung)
- 101078069/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- LCAdapt (101055309); GlycanTrigger (101093997)/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- MEM-CLONK (101055157)/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- TRR 241-A01 (375876048); SFB 1444/P11 (427826188); SPP 1937-DI764/9-2/Deutsche Forschungsgemeinschaft (German Research Foundation)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

