Molecular detection of hrHPV-induced high-grade squamous intraepithelial lesions of the cervix through a targeted RNA next generation sequencing assay
- PMID: 40448246
- PMCID: PMC12125924
- DOI: 10.1186/s10020-025-01238-x
Molecular detection of hrHPV-induced high-grade squamous intraepithelial lesions of the cervix through a targeted RNA next generation sequencing assay
Abstract
Background: Cervical cancer screening programs are increasingly relying on sensitive molecular approaches as primary tests to detect high-risk human papillomaviruses (hrHPV), the causative agents of cervix cancer. Although hrHPV infection is a pre-requisite for the development of most precancerous lesions, the mere detection of viral nucleic acids, also present in transient infections, is not specific of the underlying cellular state, resulting in poor positive predictive values (PPV) regarding lesional states. There is a need to increase the specificity of molecular tests for better stratifying individuals at risk of cancer and to adapt follow-up strategies.
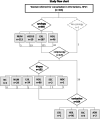
Methods: HPV-RNA-SEQ, a targeted RNA next generation sequencing assay allowing the detection of up to 16 hrHPV splice events and key human transcripts, has previously shown encouraging PPV for the detection of precancerous lesions. Herein, on 302 patients with normal cytology (NILM, n = 118), low-grade (LSIL, n = 104) or high-grade squamous intraepithelial lesions (HSIL, n = 80), machine learning-based model improvement was applied to reach 2-classes (NILM vs HSIL) or 3-classes (NILM, LSIL, HSIL) predictive models.
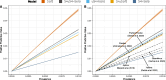
Results: Linear (elastic net) and nonlinear (random forest) approaches resulted in five 2-class models that detect HSIL vs NILM in a validation set with specificity up to 0.87, well within the range of PPV of other competing RNA-based tests in a screening population.
Conclusions: HPV-RNA-SEQ improves the detection of HSIL lesions and has the potential to complete and eventually replace current molecular approaches as a first-line test. Further performance evaluation remains to be done on larger and prospective cohorts.
Keywords: Human Papillomavirus (HPV); Molecular test; Next-Generation Sequencing (NGS); Precancerous lesions; Screening; Transcriptome.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Cervical smears were collected in the context of routine health care by the Biobanque de Picardie (BRIF N BB-0033–00017) at the CHU Amiens-Picardie. Leftover samples were used secondarily for research purposes. No additional samples were taken for this study. The biobank guarantees that the people from whom the biological samples and data came have been informed of the research and on their right of opposition, access or rectification, and have not expressed their opposition to the reuse of their biological samples and their personal data. The processing of personal data follows the rules of the European General Data Protection Regulation (GDPR). A Data Protection Officer (DPO) has been designated for this research. Information about the study has been published on the Health Data Hub ( https://www.health-data-hub.fr/ ) under reference F20220616163142. Consent for publication: Participants from whom the biological samples and data came have been informed of the research and on their right of opposition, access or rectification, and have not expressed their opposition to the reuse of their biological samples and their personal data. The processing of personal data follows the rules of the European General Data Protection Regulation (GDPR, Regulation (EU) 2016/679). Competing interests: The authors declare no competing interests.
Figures



References
-
- Cerasuolo A, Buonaguro L, Buonaguro FM, Tornesello ML. The role of RNA splicing factors in cancer: regulation of viral and human gene expression in human papillomavirus-related cervical cancer. Front Cell Dev Biol. 2020;8. Available from: https://www.frontiersin.org/articles/10.3389/fcell.2020.00474. Cited 2024 Jan 23. - DOI - PMC - PubMed

