Diverse modes of ceftazidime/avibactam resistance acquisition in carbapenem-resistant Klebsiella pneumoniae and Pseudomonas aeruginosa from a Chinese intensive care unit
- PMID: 40448249
- PMCID: PMC12125794
- DOI: 10.1186/s12941-025-00800-z
Diverse modes of ceftazidime/avibactam resistance acquisition in carbapenem-resistant Klebsiella pneumoniae and Pseudomonas aeruginosa from a Chinese intensive care unit
Abstract
Objectives: To investigate the mechanisms of ceftazidime/avibactam (CZA) resistance and the nosocomial dissemination of carbapenem-resistant Pseudomonas aeruginosa (CRPA) and carbapenem-resistant Klebsiella pneumoniae (CRKP) in an intensive care unit (ICU) in China.
Methods: Clinical CRPA and CRKP isolates were obtained from an ICU of a tertiary hospital in China from August 2020 to February 2021. Antimicrobial susceptibility was determined according to CLSI. WGS, cloning experiments and kinetic parameters were conducted to reveal resistance mechanisms, molecular characteristics and dissemination of CRPA and CRKP.
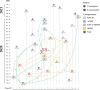
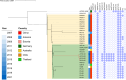
Results: We isolated 32 CZA-resistant strains, including 12 CRPA and 20 CRKP strains from an ICU between August 2020 and February 2021. CZA resistance was associated with the presence of NDM and efflux pumps in CRKP strains, whereas blaAFM-2, blaKPC-87, and blaPER-1 contributed to CZA resistance in CRPA strains. Compared to KPC-2, KPC-87 exhibited a 1.5-fold elevation in kcat/Km for ceftazidime, a 7.5-fold increase in Ki for avibactam, and a loss of carbapenem hydrolysis. blaKPC-87 was located in the NTEKPC-IIa like element based on the Tn3. Insertion of 656 bp ΔblaTEM-1 upstream of blaKPC-87 introduced an additional promoter that increased KPC-87 expression. Cluster 2 and 3 of CRKP represented two different clones of ST11 transmitted between patients. KPC-87-producing ST270 CRPA strains exhibited a small-scale dissemination and cross-regional transfer with the referral of a patient. The evolutionary pathways of AFM-2-producing ST275 CRPA strains were more complex to elucidate the transmission events.
Conclusions: In CRKP and CRPA, diverse resistance mechanisms contributed to CZA resistance. These CZA-resistant strains were transmitted among patients in the ICU and even across regions to the other healthcare unit when the patient was transferred.
Keywords: Klebsiella pneumoniae; Pseudomonas aeruginosa; Ceftazidime/avibactam; Cross-regional transmission; KPC; Non-Tn4401 elements.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing of interests: The authors declare no competing interests. Ethics statement: Approval was obtained from the Ethics Committee of Sir Run Run Shaw Hospital (approval/reference number: 20200831-36). Transparency declarations: None to declare.
Figures

Similar articles
-
Carbapenem-resistant Klebsiella pneumoniae infections in Chinese children: in vitro activities of ceftazidime-avibactam and aztreonam-avibactam against carbapenemase-producing strains in a two-center study.Front Cell Infect Microbiol. 2025 Mar 26;15:1545999. doi: 10.3389/fcimb.2025.1545999. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40207055 Free PMC article.
-
Overexpression of KPC contributes to ceftazidime-avibactam heteroresistance in clinical isolates of carbapenem-resistant Klebsiella pneumoniae.Front Cell Infect Microbiol. 2024 Dec 6;14:1450530. doi: 10.3389/fcimb.2024.1450530. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39711783 Free PMC article.
-
Molecular Mechanisms Driving the In Vivo Development of KPC-71-Mediated Resistance to Ceftazidime-Avibactam during Treatment of Carbapenem-Resistant Klebsiella pneumoniae Infections.mSphere. 2021 Dec 22;6(6):e0085921. doi: 10.1128/mSphere.00859-21. Epub 2021 Dec 22. mSphere. 2021. PMID: 34935448 Free PMC article.
-
Efficacy and safety of ceftazidime-avibactam compared to other antimicrobials for the treatment of infections caused by carbapenem-resistant Klebsiella pneumoniae strains, a systematic review and meta-analysis.Microb Pathog. 2023 Jun;179:106090. doi: 10.1016/j.micpath.2023.106090. Epub 2023 Mar 31. Microb Pathog. 2023. PMID: 37004964
-
The Hospital Water Environment as a Reservoir for Carbapenem-Resistant Organisms Causing Hospital-Acquired Infections-A Systematic Review of the Literature.Clin Infect Dis. 2017 May 15;64(10):1435-1444. doi: 10.1093/cid/cix132. Clin Infect Dis. 2017. PMID: 28200000
References
-
- Zhang P, Wang J, Li Y, Shi W, Cai H, Yang Q, Li X, Yu Y, Qu T, Jiang Y. Emergence of blaKPC-33-harboring hypervirulent ST463 Pseudomonas aeruginosa causing fatal infections in China. J Infect. 2022;85:e86–8. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

