Timed chromatin invasion during mitosis governs prototype foamy virus integration site selection and infectivity
- PMID: 40448500
- PMCID: PMC12125541
- DOI: 10.1093/nar/gkaf449
Timed chromatin invasion during mitosis governs prototype foamy virus integration site selection and infectivity
Abstract
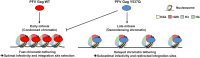
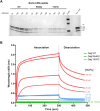
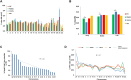
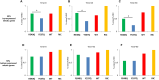
Selection of a suitable chromatin environment during retroviral integration is a tightly regulated process. Most retroviruses, including spumaretroviruses, require mitosis for nuclear entry. However, whether intrinsic chromatin dynamics during mitosis modulates retroviral genome invasion is unknown. Previous work uncovered critical interactions of prototype foamy virus (PFV) Gag with nucleosomes via a highly conserved arginine anchor residue. Yet, the regulation of Gag-chromatin interaction and its functional consequences for spumaretrovirus biology remain obscure. Here, we investigated the kinetics of chromatin binding by Gag during mitosis and proviral integration in synchronized cells. We showed that alteration of Gag affinity for nucleosome binding induced untimely chromatin tethering during mitosis, decreased infectivity, and redistributed viral integration sites to markers associated with late replication timing of chromosomes. Mutant Gag proteins were, moreover, defective in their ability to displace the histone H4 tail from the nucleosome acidic patch of highly condensed chromatin. These data indicate that the chromatin landscape during Gag-nucleosome interactions is important for PFV integration site selection and that spumaretroviruses evolved high-affinity chromatin binding to overcome early mitosis chromatin condensation.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
None declared.
Figures





References
-
- Buseyne F, Betsem E, Montange T et al. Clinical signs and blood test results among humans infected with zoonotic simian foamy virus: a case-control study. J Infect Dis. 2018; 218:144–51. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

