P4PP: A Universal Shotgun Proteomics Data Analysis Pipeline for Virus Identification
- PMID: 40449796
- PMCID: PMC12418414
- DOI: 10.1016/j.mcpro.2025.101004
P4PP: A Universal Shotgun Proteomics Data Analysis Pipeline for Virus Identification
Abstract
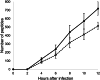
Humans can be infected by a wide variety of virus species. We developed a data analysis approach for shotgun proteomic data to detect these viruses. A proteome for pandemic preparedness (P4PP) pipeline, a corresponding database (P4PP v01), and a web application (P4PP) were constructed. The P4PP pipeline enables the identification of 1896 virus species from the 32 virus families, based on multiple identified discriminatory peptides, in which at least one human infectious virus is described. P4PP was evaluated using different datasets of cell-cultivated viruses, generated at different institutes, measured with different instruments, and prepared with different sample preparation methods. In total, 174 mass spectrometry datasets of 160 and 14 protein trypsin digests of virus-infected and noninfected cell lines were analyzed, respectively. Of the 160 samples, 146 were correctly identified at the species level, and an additional four samples were identified at the family level. In the remaining 10 samples, no virus was detected. However, all these 10 samples tested positive in follow-up samples obtained later in time series were negative samples were measured, indicating that the number of peptides derived from the virus was initially too low in the samples obtained at the start of the experiment. Furthermore, results show that influenza A or severe acute respiratory syndrome coronavirus 2 can be subtyped if enough discriminative peptides of the virus are identified. In the noninfected cell lines, no virus was detected except in one sample where the in that experiment studied virus was detected. Shotgun proteomics, in combination with the developed data analysis approach, can identify all types of virus species after cultivation in a cell line. Implementing this agnostic virus proteome analysis capability in viral diagnostic laboratories has the potential to improve their capabilities to cope with unexpected, mutated, or re-emerging viruses.
Keywords: P4PP pipeline; pandemic preparedness; peptides; proteomics; virus identification.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of Interest The authors declare no competing interests.
Figures











References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

