Precision multiplexed base editing in human cells using Cas12a-derived base editors
- PMID: 40449999
- PMCID: PMC12126522
- DOI: 10.1038/s41467-025-59653-x
Precision multiplexed base editing in human cells using Cas12a-derived base editors
Abstract
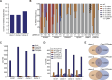
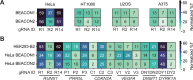
Base editors enable the direct conversion of target nucleotides without introducing DNA double strand breaks, making them a powerful tool for creating point mutations in a human genome. However, current Cas9-derived base editing technologies have limited ability to simultaneously edit multiple loci with base-pair level precision, hindering the generation of polygenic phenotypes. Here, we test the ability of six Cas12a-derived base editing systems to process multiple gRNAs from a single transcript. We identify base editor variants capable of multiplexed base editing and improve the design of the respective gRNA array expression cassette, enabling multiplexed editing of 15 target sites in multiple human cell lines, increasing state-of-the-art in multiplexing by three-fold in the field of mammalian genome engineering. To reduce bystander mutations, we also develop a Cas12a gRNA engineering approach that directs editing outcomes towards a single base-pair conversion. We combine these advances to demonstrate that both strategies can be combined to drive multiplex base editing with greater precision and reduced bystander mutation rates. Overcoming these key obstacles of mammalian genome engineering technologies will be critical for their use in studying single nucleotide variant-associated diseases and engineering synthetic mammalian genomes.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

