Genetic architecture of ideotype-related traits in middle American beans through single trait, multi-trait and epistatic genome-wide analyses
- PMID: 40450629
- PMCID: PMC12127234
- DOI: 10.1007/s00122-025-04924-w
Genetic architecture of ideotype-related traits in middle American beans through single trait, multi-trait and epistatic genome-wide analyses
Abstract
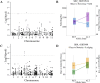
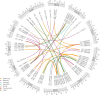
Common bean is one of the major legume crops for direct human consumption. The genetic improvement of common bean is a primary approach to increase crop adaptability to climate change conditions while maintaining productivity. A diverse panel of middle American beans was evaluated over three growing seasons (2021, 2022 and 2023) for agronomic traits considered in the crop ideotype such as flowering, maturity, pigment content, lodging, and yield. A study of the genetic regions controlling trait variation was carried out using single-trait and simultaneous (joint) multi-trait GWAS approaches. Additionally, genome-wide epistatic interactions were also analyzed. Several previously reported and novel regions were identified as significant for individual traits in the single and multi-year analyses with varying percentages of individual (7-52%) and collective (10 - 59%) phenotypic variance explained. In the single-trait and multi-year analyses, markers detected for lodging showed the highest average of percentage of variance explained (52%) followed by other traits with percentages between 11 and 19%. For yield, new loci were found with estimated effects between -96.19 to 90.96 kg/Ha in the multi-year data. In the multi-trait analyses, marginal loci on Pv02 and another on Pv04 were identified to have interaction effects on flowering and yield. A significant locus on Pv04 showed a common effect between lodging and maturity, characterized by several SNPs. Significant epistatic interactions were found along different chromosomes for all the evaluated traits, with some loci having interactions with multiple regions. In flowering, an interaction between loci on Pv01 and Pv04 explained up to 10.5% of the phenotypic variation, followed by interactions between Pv06 and Pv10 for chlorophyll b and between Pv03 and Pv08 for yield explaining around 6% of the trait variation. Multiple transcription factors were identified as candidate genes, particularly in the pairs of combinations of epistatic effects. Based on the homology analyses of the candidate genes, several showed potential roles in the genetic control of the agronomic traits, particularly for flowering, maturity, and yield. Our results demonstrate the applicability of various approaches for common bean and show a comprehensive and expanded panorama of the genetic basis of agronomic traits. The results provide new resources for crop improvement that can be leveraged in multiple approaches such as selection, modeling and predicting crop performance and genetic gain.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Conflict of interest: On behalf of all authors, the corresponding author states that there is no conflict of interest.
Figures


Similar articles
-
Multi-locus genome-wide association mapping for major agronomic and yield-related traits in sorghum (Sorghum bicolor (L.) moench) landraces.BMC Genomics. 2025 Mar 28;26(1):304. doi: 10.1186/s12864-025-11458-4. BMC Genomics. 2025. PMID: 40155810 Free PMC article.
-
Genetic mapping for agronomic traits in a MAGIC population of common bean (Phaseolus vulgaris L.) under drought conditions.BMC Genomics. 2020 Nov 16;21(1):799. doi: 10.1186/s12864-020-07213-6. BMC Genomics. 2020. PMID: 33198642 Free PMC article.
-
Genome-Wide Association Study for Major Biofuel Traits in Sorghum Using Minicore Collection.Protein Pept Lett. 2021;28(8):909-928. doi: 10.2174/0929866528666210215141243. Protein Pept Lett. 2021. PMID: 33588716
-
Genome Wide Single Locus Single Trait, Multi-Locus and Multi-Trait Association Mapping for Some Important Agronomic Traits in Common Wheat (T. aestivum L.).PLoS One. 2016 Jul 21;11(7):e0159343. doi: 10.1371/journal.pone.0159343. eCollection 2016. PLoS One. 2016. PMID: 27441835 Free PMC article.
-
Marker-based linkage map of Andean common bean (Phaseolus vulgaris L.) and mapping of QTLs underlying popping ability traits.BMC Plant Biol. 2012 Aug 9;12:136. doi: 10.1186/1471-2229-12-136. BMC Plant Biol. 2012. PMID: 22873566 Free PMC article.
References
-
- Acquaah G, Adams MW, Kelly JD (1991) Identification of effective indicators of erect plant architecture in dry bean. Crop Sci 31(2):261–264. 10.2135/CROPSCI1991.0011183X003100020004X
-
- Adams MW (1982) Plant architecture and yield breeding in Phaseolus vulgaris L. Iowa State J Res 56:225–254
-
- Alvarado GFM, Rodríguez A, Pacheco J, Burgueño J, Crossa M, Vargas P-P-R, Lopez-Cruz MA (2020) META-R: a software to analyze data from multi-environment plant breeding trials. Crop J 8:745–756. 10.1016/j.cj.2020.03.010
-
- Bates D, Mächler M, Bolker BM, Walker SC (2015) Fitting linear mixed-effects models using lme4. J Stat Softw 67:1–48. 10.18637/jss.v067.i01
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

