An attempt to explain what intrinsically disordered TCF4 does in its spare time when PTHS-related mutations prevent it from doing its job
- PMID: 40452023
- PMCID: PMC12128249
- DOI: 10.1186/s12964-025-02265-1
An attempt to explain what intrinsically disordered TCF4 does in its spare time when PTHS-related mutations prevent it from doing its job
Abstract
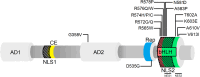
Pitt-Hopkins Syndrome (PTHS) is a rare neurodevelopmental disorder caused by mutations in the TCF4 gene (18q21.2), encoding the transcription factor 4 (TCF4). This protein is critical for central nervous system development and neuronal maturation. Mutations in TCF4, which range from point mutations to large deletions, result in varying clinical severity, including intellectual disability (ID), motor impairments, and autistic features. Despite its rarity, PTHS has gained increasing attention due to advances in understanding the genetic and molecular mechanisms underlying TCF4 function. Recent research has enhanced diagnostic approaches, including genetic testing techniques like genome sequencing, enabling more accurate identification of the disorder. Despite the evident enhancement in the PTHS management from a medical standpoint, the molecular underpinnings of the disorder progression remain puzzling. This is particularly the case where the disease is caused by point mutations. This review summarizes the latest findings on TCF4 function in PTHS, discusses the variability in mutation effects, highlights current diagnostic and therapeutic advancements, and attempts to explain the molecular bases of mutated TCF4 malfunctionality.
Keywords: Genetic disorder; Neurodevelopment; Pitt-Hopkins syndrome (PTHS); Protein aggregation; Rare disease; Transcription factor 4 (TCF4).
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
Common Pathophysiology in Multiple Mouse Models of Pitt-Hopkins Syndrome.J Neurosci. 2018 Jan 24;38(4):918-936. doi: 10.1523/JNEUROSCI.1305-17.2017. Epub 2017 Dec 8. J Neurosci. 2018. PMID: 29222403 Free PMC article.
-
Impairment of different protein domains causes variable clinical presentation within Pitt-Hopkins syndrome and suggests intragenic molecular syndromology of TCF4.Eur J Med Genet. 2017 Nov;60(11):565-571. doi: 10.1016/j.ejmg.2017.08.004. Epub 2017 Aug 12. Eur J Med Genet. 2017. PMID: 28807867
-
Disordered breathing in a Pitt-Hopkins syndrome model involves Phox2b-expressing parafacial neurons and aberrant Nav1.8 expression.Nat Commun. 2021 Oct 13;12(1):5962. doi: 10.1038/s41467-021-26263-2. Nat Commun. 2021. PMID: 34645823 Free PMC article.
-
Pitt-Hopkins Syndrome: intellectual disability due to loss of TCF4-regulated gene transcription.Exp Mol Med. 2013 May 3;45(5):e21. doi: 10.1038/emm.2013.32. Exp Mol Med. 2013. PMID: 23640545 Free PMC article. Review.
-
Molecular and Cellular Function of Transcription Factor 4 in Pitt-Hopkins Syndrome.Dev Neurosci. 2021;43(3-4):159-167. doi: 10.1159/000516666. Epub 2021 Jun 16. Dev Neurosci. 2021. PMID: 34134113 Free PMC article. Review.
References
-
- Pitt D, Hopkins IA. Syndrome of mental retardation, wide mouth and intermittent overbreathing. Aust Paediatr J. 1978. 10.1111/jpc.1978.14.3.182. - PubMed
-
- Sweetser DA, Elsharkawi I, Yonker L, Steeves M, Parkin K, Thibert R, Pitt-Hopkins S, Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Amemiya. A., Eds.; 2018.
-
- Giurgea I, Missirian C, Cacciagli P, Whalen S, Fredriksen T, Gaillon T, Rankin J, Mathieu-Dramard M, Morin G, Martin-Coignard D, et al. TCF4 deletions in Pitt-Hopkins syndrome. Hum Mutat. 2008;29:242–51. 10.1002/humu.20859. - PubMed
-
- Stavropoulos DJ, MacGregor DL, Yoon G. Mosaic microdeletion 18q21 as a cause of mental retardation. Eur J Med Genet. 2010;53:396–9. 10.1016/j.ejmg.2010.08.005. - PubMed
-
- Rossi M, Labalme A, Cordier MP, Till M, Blanchard G, Dubois R, Guibaud L, Heissat S, Javouhey E, Lachaux A, et al. Mosaic 18q21.2 deletions including the TCF4 gene: A clinical report. Am J Med Genet Part A. 2012;158 A:3174–81. 10.1002/ajmg.a.35588. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

