Genomic selection: Essence, applications, and prospects
- PMID: 40452138
- PMCID: PMC12127607
- DOI: 10.1002/tpg2.70053
Genomic selection: Essence, applications, and prospects
Abstract
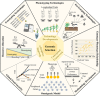
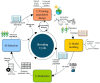
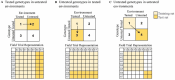
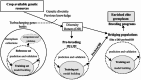
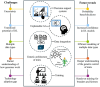
Genomic selection (GS) emerged as a key part of the solution to ensure the food supply for the growing human population thanks to advances in genotyping and other enabling technologies and improved understanding of the genotype-phenotype relationship in quantitative genetics. GS is a breeding strategy to predict the genotypic values of individuals for selection using their genotypic data and a trained model. It includes four major steps: training population design, model building, prediction, and selection. GS revises the traditional breeding process by assigning phenotyping a new role of generating data for the building of prediction models. The increased capacity of GS to evaluate more individuals, in combination with shorter breeding cycle times, has led to wide adoption in plant breeding. Research studies have been conducted to implement GS with different emphases in crop- and trait-specific applications, prediction models, design of training populations, and identifying factors influencing prediction accuracy. GS plays different roles in plant breeding such as turbocharging of gene banks, parental selection, and candidate selection at different stages of the breeding cycle. It can be enhanced by additional data types such as phenomics, transcriptomics, metabolomics, and enviromics. In light of the rapid development of artificial intelligence, GS can be further improved by either upgrading the entire framework or individual components. Technological advances, research innovations, and emerging challenges in agriculture will continue to shape the role of GS in plant breeding.
© 2025 The Author(s). The Plant Genome published by Wiley Periodicals LLC on behalf of Crop Science Society of America.
Conflict of interest statement
Patrick S. Schnable is a co‐founder and CEO of Dryland Genetics, Inc and a co‐founder and managing partner of Data2Bio, LLC and EnGeniousAg, LLC. He is a member of the scientific advisory boards of Kemin Industries and Centro de Tecnologia Canavieira. He is a recipient of research funding from Iowa Corn and Bayer Crop Science. The other authors declare no conflicts of interest.
Figures
References
-
- Abdollahi‐Arpanahi, R. , Morota, G. , Valente, B. D. , Kranis, A. , Rosa, G. J. M. , & Gianola, D. (2016). Differential contribution of genomic regions to marked genetic variation and prediction of quantitative traits in broiler chickens. Genetics, Selection, Evolution, 48(10), 10. 10.1186/s12711-016-0187-z - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- 2021-67013-33833/National Institute of Food and Agriculture
- 2023-70412-41087/National Institute of Food and Agriculture
- Hatch project (1021013)/National Institute of Food and Agriculture
- In-House Project 2090-21000-033-00D/Agricultural Research Service
- In-House Project 5030-21000-065-000-D/Agricultural Research Service
LinkOut - more resources
Full Text Sources

