EDS-Kcr: deep supervision based on large language model for identifying protein lysine crotonylation sites across multiple species
- PMID: 40452145
- PMCID: PMC12127148
- DOI: 10.1093/bib/bbaf249
EDS-Kcr: deep supervision based on large language model for identifying protein lysine crotonylation sites across multiple species
Abstract
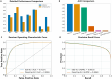
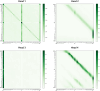
With the rapid advancement of proteomics, post-translational modifications, particularly lysine crotonylation (Kcr), have gained significant attention in basic research, drug development, and disease treatment. However, current methods for identifying these modifications are often complex, costly, and time-consuming. To address these challenges, we have proposed EDS-Kcr, a novel bioinformatics tool that integrates the state-of-the-art protein language model ESM2 with deep supervision to improve the efficiency and accuracy of Kcr site prediction. EDS-Kcr demonstrated outstanding performance across various species datasets, proving its applicability to a wide range of proteins, including those from humans, plants, animals, and microbes. Compared to existing Kcr site prediction models, our model excelled in multiple key performance indicators, showcasing superior predictive power and robustness. Furthermore, we enhanced the transparency and interpretability of EDS-Kcr through visualization techniques and attention mechanisms. In conclusion, the EDS-Kcr model provides an efficient and reliable predictive tool suitable for disease diagnosis and drug development. We have also established a freely accessible web server for EDS-Kcr at http://eds-kcr.lin-group.cn/.
Keywords: deep learning model; lysine crotonylation; post-translational modifications; protein language model; web server.
© The Author(s) 2025. Published by Oxford University Press.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

