Lactate-Induced Mitochondrial Calcium Uptake 3 Aggravates Myocardial Ischemia-Reperfusion Injury by Promoting Neutrophil Extracellular Trap Formation
- PMID: 40452820
- PMCID: PMC12123085
- DOI: 10.34133/research.0705
Lactate-Induced Mitochondrial Calcium Uptake 3 Aggravates Myocardial Ischemia-Reperfusion Injury by Promoting Neutrophil Extracellular Trap Formation
Abstract
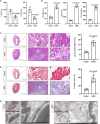
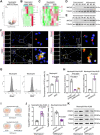
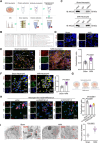
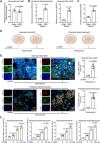
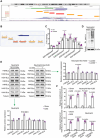
Background: Ischemic heart disease is a leading cause of mortality and disability worldwide among cardiovascular conditions. Myocardial ischemia-reperfusion injury (MIRI) occurs following percutaneous coronary intervention, during which neutrophils generate neutrophil extracellular traps (NETs) in response to injury. This study aims to elucidate the mechanisms underlying NET activation and its impact on MIRI. Methods: Sham and MIRI rat models were established. Various techniques, including enzyme-linked immunosorbent assay, hematoxylin and eosin staining, Masson staining, and transmission electron microscopy, were used to assess endothelial cell injury and myocardial tissue inflammation. Immunofluorescence was employed to evaluate NET activation in tissues, peripheral blood neutrophils, and protein colocalization. MitoTracker and ER-Tracker staining were conducted to assess the formation of mitochondria-associated membranes (MAMs). Extracted NETs were applied to conduct microvascular endothelial cell tube formation assay and flow cytometry. RNA-sequencing and immunoprecipitation-mass spectrometry were applied to determine the key regulators. Flow cytometry and Western blot were used to assess Ca2+ and mitophagy levels in neutrophils. Deoxyribonuclease I, NET inhibitor, was injected into MIRI rats to evaluate the in vivo effects of NET modulation on MIRI severity. Results: MIRI was often accompanied by cardiac microvascular endothelial cell (CMEC) injury and inflammation. Lactate mediated H3K18 lactylation at the MICU3 promoter in neutrophils, enhancing its transcription and leading to elevated MICU3 levels. Besides, lactate also promoted the interaction between MICU3 and AASR1, stabilizing MICU3 through lactylation. Up-regulated MICU3 interacted with VDAC1, facilitating MAM formation, excessive Ca2+ uptake, mitochondrial dysfunction, mitophagy activation, and NET activation. Elevated NET level exacerbated CMEC dysfunction, further aggravating MIRI. Conclusion: Lactate-driven MICU3 transcriptional activation and stabilization facilitates NET formation, contributing to MIRI development.
Copyright © 2025 Hongru Zhang et al.
Conflict of interest statement
Author contributions: H.Z., Y.M. designed and conducted the research. S.X., L.L., and C.S. performed the experiments. X.J., J.L. analyzed the results. J.C. wrote the paper. Competing interests: The authors declare that they have no competing interests.
Figures



References
-
- Guan C, Wu S, Xu W, Zhang J. Global, regional, and national burden of ischaemic heart disease and its trends, 1990-2019. Public Health. 2023;223:57–66. - PubMed
-
- Crea F, Montone RA, Rinaldi R. Pathophysiology of coronary microvascular dysfunction. Circ J. 2022;86(9):1319–1328. - PubMed
-
- Marzilli M, Merz CNB, Boden WE, Bonow RO, Capozza PG, Chilian WM, DeMaria AN, Guarini G, Huqi A, Morrone D, et al. Obstructive coronary atherosclerosis and ischemic heart disease: An elusive link! J Am Coll Cardiol. 2012;60(11):951–956. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

