A feedforward loop between ACLY and MYC supports T-ALL progression in vivo
- PMID: 40453140
- PMCID: PMC12067888
- DOI: 10.1016/j.bneo.2025.100069
A feedforward loop between ACLY and MYC supports T-ALL progression in vivo
Abstract
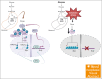
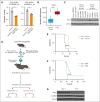
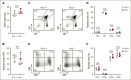
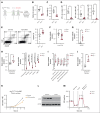
T-cell acute lymphoblastic leukemia (T-ALL) is a hematological malignancy in need of novel therapeutic approaches. Here, we identify ATP-citrate lyase (ACLY) as overexpressed in human T-ALL and as a promising therapeutic target for its treatment. To test the effects of ACLY in leukemia progression, we developed an isogenic model of NOTCH1-induced Acly conditional knockout leukemia. Importantly, we observed intrinsic antileukemic effects upon loss of ACLY, which further synergized with NOTCH1 inhibition in vivo. Metabolomic profiling upon ACLY loss revealed a metabolic crisis with reduced acetyl-coenzyme A (acetyl-CoA) levels and decreased oxygen consumption rate. Gene expression profiling analyses showed that the transcriptional signature of ACLY loss very significantly correlates with the signature of MYC loss in vivo. Mechanistically, the decrease in acetyl-CoA led to reduced H3K27ac levels in Myc, resulting in transcriptional downregulation of Myc and drastically reduced MYC protein levels. Moreover, pharmacological inhibition of ACLY led to reduced MYC levels and antileukemic effects in human T-ALL cell lines and patient-derived xenografts. Interestingly, our analyses also revealed a reciprocal relationship whereby ACLY itself is a direct transcriptional target of MYC, thus establishing a feedforward loop that is important for leukemia progression. Overall, our results identified a relevant ACLY-MYC axis and unveiled ACLY as a novel promising target for T-ALL treatment.
© 2025 American Society of Hematology. Published by Elsevier Inc. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures

References
-
- Pagliaro L, Chen SJ, Herranz D, et al. Acute lymphoblastic leukaemia. Nat Rev Dis Primers. 2024;10(1):41. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

