Identifying Common Diagnostic Biomarkers and Therapeutic Targets between COPD and Sepsis: A Bioinformatics and Machine Learning Approach
- PMID: 40453984
- PMCID: PMC12126980
- DOI: 10.2147/COPD.S510846
Identifying Common Diagnostic Biomarkers and Therapeutic Targets between COPD and Sepsis: A Bioinformatics and Machine Learning Approach
Abstract
Background: Evidence suggests a bidirectional association between chronic obstructive pulmonary disease (COPD) and sepsis, but the underlying mechanisms remain unclear. This study aimed to explore shared diagnostic genes, potential mechanisms, and the role of immune cells in the COPD-sepsis relationship using Mendelian randomization (MR) and bioinformatics approaches, while also identifying potential therapeutic drugs.
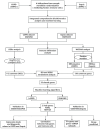
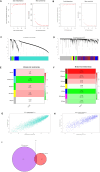
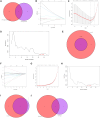
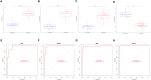
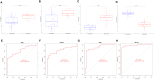
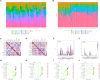
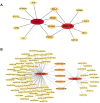
Methods: Two-sample MR analysis was performed using genome-wide association data to assess genetically predicted COPD and sepsis. Immune cell-mediated effects were quantified using a two-way two-sample MR analysis. Differential expression gene (DEG) analysis and weighted gene co-expression network analysis (WGCNA) were used to identify common genes. Functional enrichment analyses were conducted to explore the biological roles of these genes. LASSO and SVM-RFE algorithms identified shared diagnostic genes, which were evaluated using receiver operating characteristic (ROC) curves. Immune cell infiltration was analyzed with CIBERSORT, while transcription factor (TF) and miRNA networks were constructed using NetworkAnalyst. Drug predictions were made using DSigDB, and molecular docking validated potential drugs.
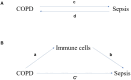
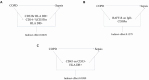
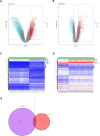
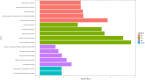
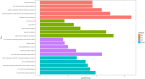
Results: Three immune cell types were identified as mediators between COPD and sepsis, with genetically predicted effects mediated by these cells at rates of 6.5%, 12.8%, and 3.9%. A total of 33 overlapping genes were identified, and AIM2 and RNF125 were highlighted as key diagnostic genes. Immune infiltration analysis revealed dysregulated monocyte, macrophage, plasma, and dendritic cells. Regulatory network analysis identified nine key co-regulators. Ten potential drug targets were identified, with seven validated via molecular docking.
Conclusion: AIM2 and RNF125 may serve as diagnostic biomarkers, and identified immune cell subsets could mediate the COPD-sepsis connection, offering insights into potential therapeutic targets.
Keywords: Mendelian randomization; chronic obstructive pulmonary disease; co-diagnostic genes; comprehensive bioinformatics analysis; immune cells; machine learning; molecular docking; predictive drugs; sepsis.
© 2025 Li et al.
Conflict of interest statement
Xinyi Li and Yuyang Xiao are co-first authors for this study. The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

