Gold Nanoparticle-DNA Conjugates: An Enzymatic DNA Synthesis Platform
- PMID: 40454042
- PMCID: PMC12120612
- DOI: 10.1021/acsomega.5c00645
Gold Nanoparticle-DNA Conjugates: An Enzymatic DNA Synthesis Platform
Abstract
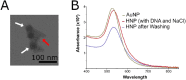
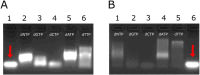
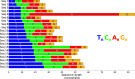
Leveraging the well-established properties of gold nanoparticle (AuNP)-DNA conjugates, this research explored a novel methodology for controlled enzymatic DNA synthesis using gold nanoparticle (AuNP)-DNA conjugates as a solid platform. To this end, Hybrid Nanoparticles (HNPs) were meticulously engineered through the functionalization of AuNPs with rationally designed DNA initiator molecules. These initiator molecules, strategically attached to the AuNP surface, served as a physical support and starting point for DNA extension by the Terminal Deoxynucleotidyl Transferase (TdT) enzyme. The results confirmed the synthesis of homopolymeric DNA extensions on these HNPs (58.27 nm, PDI < 0.2), demonstrating the viability of HNPs as a platform for enzymatic DNA elongation. Although the growing demand for data storage suggests a potential application, this research established the foundational feasibility of enzymatic DNA synthesis on HNPs. While high-density DNA data storage requires extensive development, the demonstrated enzymatic synthesis on AuNP-DNA conjugates warrants significant further exploration for future applications in biotechnology and nanotechnology.
© 2025 The Authors. Published by American Chemical Society.
Figures




Similar articles
-
Stable gold nanoparticle conjugation to internal DNA positions: facile generation of discrete gold nanoparticle-DNA assemblies.Bioconjug Chem. 2010 Aug 18;21(8):1413-6. doi: 10.1021/bc100160k. Bioconjug Chem. 2010. PMID: 20666441
-
Spectroscopic studies under properties of chlorpromazine conjugated to gold nanoparticles.Spectrochim Acta A Mol Biomol Spectrosc. 2024 Nov 5;320:124588. doi: 10.1016/j.saa.2024.124588. Epub 2024 Jun 3. Spectrochim Acta A Mol Biomol Spectrosc. 2024. PMID: 38870699
-
Applications of Terminal Deoxynucleotidyl Transferase Enzyme in Biotechnology.Chembiochem. 2023 Mar 1;24(5):e202200510. doi: 10.1002/cbic.202200510. Epub 2022 Nov 24. Chembiochem. 2023. PMID: 36342345 Review.
-
An ultrasensitive signal-on electrochemical aptasensor for ochratoxin A determination based on DNA controlled layer-by-layer assembly of dual gold nanoparticle conjugates.Biosens Bioelectron. 2018 Oct 15;117:845-851. doi: 10.1016/j.bios.2018.07.012. Epub 2018 Jul 11. Biosens Bioelectron. 2018. PMID: 30096739
-
Design and applications of gold nanoparticle conjugates by exploiting biomolecule-gold nanoparticle interactions.Nanoscale. 2013 Apr 7;5(7):2589-99. doi: 10.1039/c3nr33870c. Nanoscale. 2013. PMID: 23423633 Review.
Cited by
-
From Past to Present: Gold Nanoparticles (AuNPs) in Daily LifeSynthesis Mechanisms, Influencing Factors, Characterization, Toxicity, and Emerging Applications in Biomedicine, Nanoelectronics, and Materials Science.ACS Omega. 2025 Jul 30;10(31):33999-34087. doi: 10.1021/acsomega.5c03162. eCollection 2025 Aug 12. ACS Omega. 2025. PMID: 40821590 Free PMC article. Review.
References
-
- Bögels B. W. A., Nguyen B. H., Ward D., Gascoigne L., Schrijver D. P., Makri Pistikou A.-M., Joesaar A., Yang S., Voets I. K., Mulder W. J. M., Phillips A., Mann S., Seelig G., Strauss K., Chen Y.-J., de Greef T. F. A.. DNA Storage in Thermoresponsive Microcapsules for Repeated Random Multiplexed Data Access. Nat. Nanotechnol. 2023;18(8):912–921. doi: 10.1038/s41565-023-01377-4. - DOI - PMC - PubMed
-
- Doricchi A., Platnich C. M., Gimpel A., Horn F., Earle M., Lanzavecchia G., Cortajarena A. L., Liz-Marzán L. M., Liu N., Heckel R., Grass R. N., Krahne R., Keyser U. F., Garoli D.. Emerging Approaches to DNA Data Storage: Challenges and Prospects. ACS Nano. 2022;16(11):17552–17571. doi: 10.1021/acsnano.2c06748. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
