D7A Mutagenesis and In Silico Analysis on Haloacid Dehalogenase Gene from Pseudomonas aeruginosa ITB1 in pET/ paed‑d Recombinant Clone
- PMID: 40454071
- PMCID: PMC12120611
- DOI: 10.1021/acsomega.4c10345
D7A Mutagenesis and In Silico Analysis on Haloacid Dehalogenase Gene from Pseudomonas aeruginosa ITB1 in pET/ paed‑d Recombinant Clone
Abstract
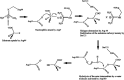
Organohalogens, including haloalkanoic acids, are organic compounds synthesized in large quantities for various purposes in the agriculture, pharmaceutics, textile, and chemical industries. Unfortunately, organohalogen waste is persistent in the environment and toxic to many organisms, including humans. In this respect, bioremediation is considered the safest and most ecofriendly technique to reduce this class of xenobiotics. Pseudomonas aeruginosa ITB1 local strain has been identified to produce haloacid dehalogenase, a hydrolytic enzyme that catalyzes the breakage of carbon-halogen covalent bonds in haloalkanoic acids. Our previous studies have successfully cloned the haloacid dehalogenase gene from this bacterium into pGEM-T Easy in E. coli TOP10, sequenced, and then subcloned into pET-30a-(+) for expression in E. coli BL21-(DE3). The gene was named paed-d, and its enzyme was called Paed-d. In silico analyses suggested that the D7 residue plays an essential role in catalysis. This study was aimed to evaluate the hypothesis by performing mutagenesis of D7A on the cloned gene using a PCR approach. The obtained amplicon was used to transform E. coli TOP10 to confirm the mutation (paed-d D7A) by sequencing and to transform E. coli BL21-(DE3) for expression analyses. Wet lab analysis demonstrated that the Paed-d D7A mutant enzyme was revealed to have 2.5-fold lowered specific activity compared to the wild type. This result proved the hypothesis that D7 in haloacid dehalogenase plays an important role in the catalysis process.
© 2025 The Authors. Published by American Chemical Society.
Figures






References
-
- Peng P., Goris T., Lu Y., Nijsse B., Burrichter A., Schleheck D., Koehorst J. J., Liu J., Sipkema D., Sinninghe Damste J. S., Stams A. J. M., Häggblom M. M., Smidt H., Atashgahi S.. Organohalide-Respiring Desulfoluna Species Isolated from Marine Environments. ISME J. 2020;14:815. doi: 10.1038/s41396-019-0573-y. - DOI - PMC - PubMed
-
- Gribble G. W.. A Recent Survey of Naturally Occurring Organohalogen Compounds. Environ. Chem. 2015;12:396. doi: 10.1071/EN15002. - DOI
-
- Gribble G. W.. Recent Discoveries of Naturally Occurring Halogenated Nitrogen Heterocycles. Prog. Heterocycl. Chem. 2021;33:1. doi: 10.1016/B978-0-323-98410-2.00001-1. - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous
