Significant underascertainment in Huntington's disease
- PMID: 40454200
- PMCID: PMC12123046
- DOI: 10.1093/braincomms/fcaf194
Significant underascertainment in Huntington's disease
Abstract
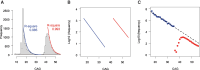
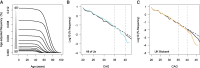
While Huntington's disease (HD), a Mendelian disorder caused by an expanded CAG repeat in HTT, is considered rare, the true prevalence could be significantly higher due to substantial underascertainment. Given inherent biases in empirically assessing disease prevalence, we performed mathematical modelling and validation analyses to estimate the frequency of expanded CAG repeats in the general population to better understand the disease prevalence. We developed an exponential decay model after confirming that the logarithmic decrease in frequency of CAG repeats extends into the pathogenic range (CAG > 35). The model was further refined by incorporating HD onset and mortality probabilities to estimate the clinical ascertainment rate. Our age-adjusted exponential decay model estimated one expanded repeat in 325 people and further showed that the frequency of expanded repeats decreases with age due to the early mortality associated with HD, which was validated by All of Us and UK Biobank data. Importantly, our data suggest that approximately half of symptomatic HD individuals aged 30-70 are not clinically ascertained/diagnosed. Our data, showing higher frequencies of expanded repeats in the general population and significant underascertainment rates, imply that HD prevalence could be twice as high as current estimates.
Keywords: Huntington’s disease; disease prevalence; exponential decay model; frequency of expanded HTT CAG repeat; underascertainment.
© The Author(s) 2025. Published by Oxford University Press on behalf of the Guarantors of Brain.
Conflict of interest statement
J.F.G. was a Scientific Advisory Board member and had a financial interest in Triplet Therapeutics, Inc. His NIH-funded project is using genetic and genomic approaches to uncover other genes that significantly influence when diagnosable symptoms emerge and how rapidly they worsen in Huntington disease. The company is developing new therapeutic approaches to address triplet repeat disorders such Huntington’s disease, myotonic dystrophy and spinocerebellar ataxias. His interests were reviewed and are managed by Massachusetts General Hospital and Mass General Brigham in accordance with their conflict of interest policies. J.F.G. has also been a consultant for Wave Life Sciences USA, Inc., Biogen, Inc. and Pfizer, Inc. J.-M.L. consults for GenKOre and serves in the scientific advisory board of GenEdit, Inc.
Figures



References
-
- HDCRG . A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell. 26 1993;72(6):971–983. - PubMed
-
- Bates GP, Dorsey R, Gusella JF, et al. Huntington disease. Nat Rev Dis Primers. 2015;1:15005. - PubMed
-
- Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. - PubMed
-
- Pearson CE, Nichol Edamura K, Cleary JD. Repeat instability: Mechanisms of dynamic mutations. Nat Rev Genet. 2005;6(10):729–742. - PubMed
LinkOut - more resources
Full Text Sources
