cfTools: an R/Bioconductor package for deconvolving cell-free DNA via methylation analysis
- PMID: 40454229
- PMCID: PMC12124914
- DOI: 10.1093/bioadv/vbaf108
cfTools: an R/Bioconductor package for deconvolving cell-free DNA via methylation analysis
Abstract
Motivation: Cell-free DNA (cfDNA) released by dying cells from damaged or diseased tissues can lead to elevated tissue-specific DNA, which is traceable and quantifiable through unique DNA methylation patterns. Therefore, tracing cfDNA origins by analyzing its methylation profiles holds great potential for detecting and monitoring a range of diseases, including cancers. However, deconvolving tissue-specific cfDNA remains challenging for broader applications and research due to the scarcity of specialized, user-friendly bioinformatics tools.
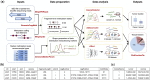
Results: To address this, we developed cfTools, an R package that streamlines cfDNA tissue-of-origin analysis for disease detection and monitoring. Integrating advanced cfDNA tissue deconvolution algorithms with R/Bioconductor compatibility, cfTools offers data preparation and analysis functions with flexible parameters for user-friendliness. By identifying abnormal cfDNA compositions, cfTools can infer the presence of underlying pathological conditions, including but not limited to cancer. It simplifies bioinformatics tasks and enables users without advanced expertise to easily derive biologically interpretable insights from standard preprocessed sequencing data, thus increasing its accessibility and broadening its application in cfDNA-based disease studies.
Availability and implementation: cfTools and its supplementary package cfToolsData are freely available at Bioconductor: https://bioconductor.org/packages/release/bioc/html/cfTools.html and https://bioconductor.org/packages/release/data/experiment/html/cfToolsData.html. The development version of cfTools is maintained on GitHub: https://github.com/jasminezhoulab/cfTools.
© The Author(s) 2025. Published by Oxford University Press.
Conflict of interest statement
X.J.Z. and W.L. are co-founders of EarlyDiagnostics, Inc. X.J.Z. serves on the Board of Directors and has an executive leadership position at EarlyDiagnostics. W.L. serves as the Board of Directors at EarlyDiagnostics. W.L. and X.J.Z. are stockholders of EarlyDiagnostics. M.L.S. is an employee of EarlyDiagnostics and has stock options with EarlyDiagnostics. W.L. and S.L. are consultants to EarlyDiagnostics and S.L. has stock options with EarlyDiagnostics. The other authors have no competing interests to declare.
Figures