PROX1 is an early driver of lineage plasticity in prostate cancer
- PMID: 40454483
- PMCID: PMC12126232
- DOI: 10.1172/JCI187490
PROX1 is an early driver of lineage plasticity in prostate cancer
Abstract
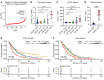
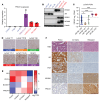
Lineage plasticity is recognized as a critical determinant of lethality and resistance to AR pathway inhibitors in prostate cancer. Lineage plasticity is a continuum, ranging from AR activity-low tumors, AR-null tumors that do not express a neuroendocrine prostate cancer (NEPC) program (i.e., double-negative prostate cancer [DNPC]), and AR-null NEPC tumors. Factors upregulated early in lineage plasticity are not well-characterized. The clarification of such factors is essential to identify tumors undergoing lineage plasticity or at risk of this occurring. Our integrative analysis of metastatic prostate cancer patient tumors, patient-derived xenografts, and cell models determined that PROX1 is upregulated early in the lineage plasticity continuum and progressively increases as tumors lose AR activity. We determined DNA methylation is a key regulator of PROX1 expression. PROX1 suppression in DNPC and NEPC reduces cell survival and impacts apoptosis and differentiation, demonstrating PROX1's functional importance. PROX1 is not directly targetable with standard drug development approaches. However, affinity immunopurification demonstrated histone deacetylases (HDACs) are among the top PROX1-interacting proteins; HDAC inhibition depletes PROX1 and recapitulates PROX1 suppression in DNPC and NEPC. Altogether, our results suggest PROX1 promotes the emergence of lineage plasticity, and HDAC inhibition is a promising approach to treat tumors across the lineage plasticity continuum.
Keywords: Cell biology; Epigenetics; Oncology; Prostate cancer.
Figures




References
MeSH terms
Substances
Grants and funding
- P50 CA275741/CA/NCI NIH HHS/United States
- P50 CA097186/CA/NCI NIH HHS/United States
- R50 CA274336/CA/NCI NIH HHS/United States
- R01 GM147365/GM/NIGMS NIH HHS/United States
- T32 GM007863/GM/NIGMS NIH HHS/United States
- R37 CA214955/CA/NCI NIH HHS/United States
- R01 CA291986/CA/NCI NIH HHS/United States
- P50 CA186786/CA/NCI NIH HHS/United States
- R01 CA282005/CA/NCI NIH HHS/United States
- T90 DE030859/DE/NIDCR NIH HHS/United States
- P01 CA163227/CA/NCI NIH HHS/United States
- P30 CA046592/CA/NCI NIH HHS/United States
- R01 CA251245/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials

