Ferroptosis-related genes mediate tumor microenvironment and prognosis in triple-negative breast cancer via integrated RNA-seq analysis
- PMID: 40454580
- PMCID: PMC12129453
- DOI: 10.7554/eLife.100923
Ferroptosis-related genes mediate tumor microenvironment and prognosis in triple-negative breast cancer via integrated RNA-seq analysis
Abstract
Triple-negative breast cancer (TNBC), an aggressive malignancy with limited tools to predict recurrence and drug sensitivity, exhibits ferroptotic heterogeneity across subtypes. However, the tumor microenvironment (TME) mediated by ferroptosis-related genes remains poorly characterized. This study integrates single-cell and bulk RNA sequencing data from the Gene Expression Omnibus to elucidate ferroptosis-driven TME features in TNBC, employing machine learning to develop prognostic and therapeutic response prediction models. At the single-cell level, T cells were classified into three subpopulations and macrophages into two subpopulations, with their infiltration degrees significantly correlated with clinical outcomes. A risk score model constructed based on these findings demonstrated robust predictive performance, validated in external cohorts with 3-, 4-, and 5-year area under the receiver operating characteristic curves of 0.65, 0.67, and 0.71, respectively. Notably, high-risk patients exhibited enhanced sensitivity to 27 therapeutic agents. By delineating ferroptosis-associated immune heterogeneity, this work provides a risk stratification tool to enhance prognostic precision and therapeutic decision-making in TNBC, while identifying genes offer actionable targets for TNBC precision medicine.
Keywords: RNA-seq; cancer biology; ferroptosis; none; triple-negative breast cancer; tumor microenvironment.
© 2024, Gong, Gu et al.
Conflict of interest statement
XG, LG, DY, YH, QL, HQ, YW No competing interests declared
Figures



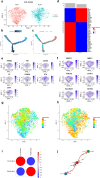


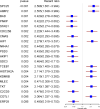



Update of
References
-
- Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine J-C, Geurts P, Aerts J, van den Oord J, Atak ZK, Wouters J, Aerts S. SCENIC: single-cell regulatory network inference and clustering. Nature Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. - DOI - PMC - PubMed
-
- Bardia A, Mayer IA, Vahdat LT, Tolaney SM, Isakoff SJ, Diamond JR, O’Shaughnessy J, Moroose RL, Santin AD, Abramson VG, Shah NC, Rugo HS, Goldenberg DM, Sweidan AM, Iannone R, Washkowitz S, Sharkey RM, Wegener WA, Kalinsky K. Sacituzumab govitecan-hziy in refractory metastatic triple-negative breast cancer. The New England Journal of Medicine. 2019;380:741–751. doi: 10.1056/NEJMoa1814213. - DOI - PubMed
-
- Bense RD, Sotiriou C, Piccart-Gebhart MJ, Haanen JBAG, van Vugt MATM, de Vries EGE, Schröder CP, Fehrmann RSN. Relevance of tumor-infiltrating immune cell composition and functionality for disease outcome in breast cancer. Journal of the National Cancer Institute. 2017;109:djw192. doi: 10.1093/jnci/djw192. - DOI - PMC - PubMed
-
- Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, Coussens LM, Gabrilovich DI, Ostrand-Rosenberg S, Hedrick CC, Vonderheide RH, Pittet MJ, Jain RK, Zou W, Howcroft TK, Woodhouse EC, Weinberg RA, Krummel MF. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nature Medicine. 2018;24:541–550. doi: 10.1038/s41591-018-0014-x. - DOI - PMC - PubMed
MeSH terms
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

