Discovery of a non-nucleoside inhibitor that binds to a novel site in the palm domain of the respiratory syncytial virus RNA-dependent RNA polymerase
- PMID: 40454903
- PMCID: PMC12282092
- DOI: 10.1128/jvi.00178-25
Discovery of a non-nucleoside inhibitor that binds to a novel site in the palm domain of the respiratory syncytial virus RNA-dependent RNA polymerase
Abstract
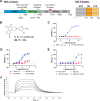
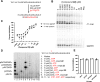
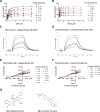
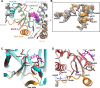
Respiratory syncytial virus (RSV) is a major cause of severe respiratory tract infections in infants, young children, and the elderly. We report herein the discovery and characterization of a novel RSV polymerase (RSVpol) non-nucleoside inhibitor (NNI) chemotype that binds to a previously undescribed, highly conserved site in the palm domain of the L protein. Consistent with the observed mode of inhibition, cryogenic electron microscopy (cryo-EM) revealed the site to be adjacent to the nucleotide binding site. Minireplicon assays confirmed on-target activity against RSVpol, and cell-based antiviral assays showed that the lead compound effectively inhibited viral mRNA transcription and replication in clinically relevant A and B strains. Together, our data provides valuable insights into the molecular basis of inhibition for a novel mechanism of action and paves the way for structure-based design to deliver effective therapeutics against RSV.IMPORTANCERespiratory syncytial virus (RSV) is a negative-sense, single-stranded RNA virus belonging to the family Pneumoviridae of the order Mononegavirales. Currently, monoclonal antibody treatments are only approved for infants, and vaccines are reserved for pregnant women and adults aged 60 years and older. Prophylaxis is also limited to the pediatric patient population, and there are currently no direct antiviral therapies for post-exposure treatment. Viral polymerases are considered well-validated drug targets due to their critical role in transcription and genome replication. Herein, we disclose the discovery of a spiro-indolinone series as polymerase inhibitors and describe the preliminary structure-activity relationship (SAR). A cryogenic electron microscopy (cryo-EM) structure obtained with an optimized lead revealed a novel binding site located in the palm domain, which will enable future structure-based drug design efforts. Novel RSV antivirals could be beneficial both as therapeutics following diagnosis and as a prophylactic in patients less likely to respond to vaccines.
Keywords: NNI; RSV polymerase inhibitor; cryogenic electron microscopy; high-throughput screen; palm domain; respiratory syncytial virus.
Conflict of interest statement
This work was funded by Johnson & Johnson Innovative Medicine. All authors were affiliated with the company at the time the work was carried out and may be Johnson & Johnson stockholders.
Figures

References
-
- Chaw PS, Wong SWL, Cunningham S, Campbell H, Mikolajczyk R, Nair H, RESCEU Investigators . 2020. Acute lower respiratory infections associated with respiratory syncytial virus in children with underlying congenital heart disease: systematic review and meta-analysis. J Infect Dis 222:S613–S619. doi: 10.1093/infdis/jiz150 - DOI - PubMed
-
- Chatzis O, Darbre S, Pasquier J, Meylan P, Manuel O, Aubert JD, Beck-Popovic M, Masouridi-Levrat S, Ansari M, Kaiser L, Posfay-Barbe KM, Asner SA. 2018. Burden of severe RSV disease among immunocompromised children and adults: a 10 year retrospective study. BMC Infect Dis 18:111. doi: 10.1186/s12879-018-3002-3 - DOI - PMC - PubMed
-
- Shi T, Denouel A, Tietjen AK, Campbell I, Moran E, Li X, Campbell H, Demont C, Nyawanda BO, Chu HY, Stoszek SK, Krishnan A, Openshaw P, Falsey AR, Nair H, RESCEU Investigators . 2020. Global disease burden estimates of respiratory syncytial virus-associated acute respiratory infection in older adults in 2015: a systematic review and meta-analysis. J Infect Dis 222:S577–S583. doi: 10.1093/infdis/jiz059 - DOI - PubMed
-
- Li Y, Johnson EK, Shi T, Campbell H, Chaves SS, Commaille-Chapus C, Dighero I, James SL, Mahé C, Ooi Y, Paget J, van Pomeren T, Viboud C, Nair H. 2021. National burden estimates of hospitalisations for acute lower respiratory infections due to respiratory syncytial virus in young children in 2019 among 58 countries: a modelling study. Lancet Respir Med 9:175–185. doi: 10.1016/S2213-2600(20)30322-2 - DOI - PubMed
-
- Chaw PS, Hua L, Cunningham S, Campbell H, Mikolajczyk R, Nair H, RESCEU Investigators . 2020. Respiratory syncytial virus-associated acute lower respiratory infections in children with bronchopulmonary dysplasia: systematic review and meta-analysis. J Infect Dis 222:S620–S627. doi: 10.1093/infdis/jiz492 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

