Concerted transport and phosphorylation of diacylglycerol at ER-PM contact sites regulate phospholipid dynamics during stress
- PMID: 40455983
- PMCID: PMC12167946
- DOI: 10.1073/pnas.2421334122
Concerted transport and phosphorylation of diacylglycerol at ER-PM contact sites regulate phospholipid dynamics during stress
Abstract
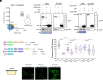
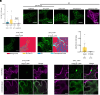
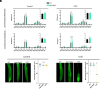
A universal response of plants to environmental stresses is the activation of plasma membrane (PM) phospholipase C, which hydrolyzes phosphoinositides to produce soluble inositol phosphate and diacylglycerol (DAG). Because of their conical shape, DAG amounts have to be tightly regulated or they can destabilize membranes. We previously showed that upon stress, Synaptotagmin1 (SYT1) transports DAG from the PM to the endoplasmic reticulum (ER) at ER-PM Contact Sites (CS). Here, we addressed the fate of the incoming DAG in the ER. We show that diacylglycerol kinases (DGKs) DGK1 and DGK2 form a module with SYT1 functionally coupling DAG transport and phosphorylation at ER-PM CS. Although SYT1 and DGK1/DGK2 do not show exclusive ER-PM CS localization, their interaction occurs specifically at ER-PM CS and the removal of ER-PM CS abolishes the interaction. Lipidomic analysis of a dgk1dgk2 double mutant supports that DGK1 and DGK2 phosphorylate DAG at the ER and transcriptomic and phenotypic analyses indicate that SYT1 and DGK1/DGK2 are functionally related. Taken together, our results highlight a mechanism at ER-PM CS that coordinates the transfer of DAG from the PM to the ER by SYT1 upon stress and the concomitant phosphorylation of DAG by DGK1 and DGK2 at the ER. These findings underscore the critical role of spatial coordination in lipid metabolism during stress-induced membrane remodeling.
Keywords: DAG; PI cycle; abiotic stress; contact sites; signaling.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures

References
-
- Jaillais Y., et al. , Guidelines for naming and studying plasma membrane domains in plants. Nat. Plants 10, 1172–1183 (2024). - PubMed
-
- Pérez-Sancho J., et al. , Stitching organelles: Organization and function of specialized membrane contact sites in plants. Trends Cell Biol. 26, 705–717 (2016). - PubMed
-
- Chang C. L., et al. , Feedback regulation of receptor-induced Ca2+ signaling mediated by E-SYT1 and NIR2 at endoplasmic reticulum-plasma membrane junctions. Cell Rep. 5, 813–825 (2013). - PubMed
MeSH terms
Substances
Grants and funding
- PID2020-114419RB-I00/AEI/10.13039/501100011033/Ministerio de Ciencia e Innovación (MCIN)
- PRE2018-085284/Ministerio de Ciencia e Innovación (MCIN)
- PID2023-147983OB-I00/Ministerio de Ciencia e Innovación (MCIN)
- PID2021-127649OB-I00/Ministerio de Ciencia e Innovación (MCIN)
- PAIDI2020-P20-00222-R/Consejería de Conocimiento, Investigación y Universidad, Junta de Andalucía (Ministry of Knowledge, Research and University, Andalusia)
- UMA20- FEDERJA-007/Consejería de Conocimiento, Investigación y Universidad, Junta de Andalucía (Ministry of Knowledge, Research and University, Andalusia)
- 101001097/EC | ERC | HORIZON EUROPE European Research Council (ERC)
- 466-2022/European Molecular Biology Organization (EMBO)
- BB/X010988/1/UKRI | Biotechnology and Biological Sciences Research Council (BBSRC)
- PID2019-106987RB-I00/Ministerio de Ciencia e Innovación (MCIN)
- PAIDI 2020-PY20_00084/Consejería de Economía, Conocimiento, Empresas y Universidad, Junta de Andalucía (Ministry of Economy, Knowledge, Business and University, Andalusia)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

