MyD88 knockdown by RNAi prevents bacterial stimulation of tubeworm metamorphosis
- PMID: 40455987
- PMCID: PMC12167997
- DOI: 10.1073/pnas.2505805122
MyD88 knockdown by RNAi prevents bacterial stimulation of tubeworm metamorphosis
Abstract
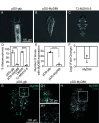
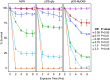
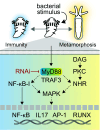
Diverse animals across the tree of life undergo the life-history transition of metamorphosis in response to bacteria. Although immunity has been implicated in this metamorphosis in response to bacteria, no functional connection has yet been demonstrated between immunity and metamorphosis. We investigated a host-microbe interaction involving a marine tubeworm, Hydroides elegans, that undergoes metamorphosis in response to Pseudoalteromonas luteoviolacea, a metamorphosis-inducing marine bacterium. By creating a marine bacteria-mediated RNA interference approach, we show that myeloid differentiation factor 88 (MyD88), a critical immune adaptor for Toll-like receptor and interleukin pathways, is necessary for the stimulation of metamorphosis in response to bacteria. In addition to a developmental role, we show that MyD88 is necessary for survival during exposure to the bacterial pathogen Pseudomonas aeruginosa, showing that Hydroides utilizes MyD88 during both development and an immune response. These results provide a functional characterization of the innate immune system involved in an animal's metamorphosis.
Keywords: MyD88; bacteria; development; immunity; metamorphosis.
Conflict of interest statement
Competing interests statement:E.D., M.V.F. and N.J.S. are co-inventors on provisional US patent application serial number 63/687,110, entitled “Genetically Engineered Marine Bacteria for Biomaterial Production, Probiotic Use in Aquaculture and Marine Environmental Restoration” and assigned to San Diego State University Research Foundation.
Figures



Comment in
-
An ancient immune pathway enables animal metamorphosis.Proc Natl Acad Sci U S A. 2025 Jul 29;122(30):e2512734122. doi: 10.1073/pnas.2512734122. Epub 2025 Jul 21. Proc Natl Acad Sci U S A. 2025. PMID: 40690684 Free PMC article. No abstract available.
References
-
- Gilbert S. F., Bosch T. C. G., Ledón-Rettig C., Eco-evo-devo: Developmental symbiosis and developmental plasticity as evolutionary agents. Nat. Rev. Genet. 16, 611–622 (2015). - PubMed
-
- Schmidt B. R., Hödl W., Schaub M., From metamorphosis to maturity in complex life cycles: Equal performance of different juvenile life history pathways. Ecology 93, 657–667 (2012). - PubMed
-
- Warkentin K. M., Environmentally cued hatching across taxa: Embryos respond to risk and opportunity. Integr. Comp. Biol. 51, 14–25 (2011). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

