Linking Virulence and Iron Limitation Response in Staphylococcus aureus: The sRNA IsrR Is Involved in SaeRS Activation
- PMID: 40456523
- PMCID: PMC12235713
- DOI: 10.1021/acs.jproteome.5c00059
Linking Virulence and Iron Limitation Response in Staphylococcus aureus: The sRNA IsrR Is Involved in SaeRS Activation
Abstract
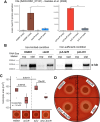
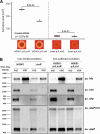
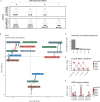
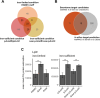
The Gram-positive opportunistic pathogen Staphylococcus aureus colonizes ∼30% of the human population but also causes various diseases. Precise regulation of genes involved in virulence and metabolic functions is required to adapt to changing host conditions, such as severe restriction of iron availability. In addition to the global regulator Fur (ferric uptake regulator), the iron limitation response of S. aureus is shaped by the recently identified sRNA IsrR (iron sparing response regulator). IsrR mediates an iron sparing response by inhibiting the synthesis of nonessential iron-containing proteins, which are in particular involved in the central metabolism. In addition, we demonstrate that isrR expression is positively associated with α-hemolysin levels and the hemolysis activity of S. aureus HG001. To investigate the influence of IsrR on virulence factor production, we performed a mass spectrometry-based secretome analysis of isrR-expressing and nonexpressing strains under iron-limited and iron-sufficient conditions. The SaeR regulon was positively influenced by the presence of IsrR, and IsrR is likely involved in the activation of the Sae system. Additionally, IsrR also positively affected the protein levels of the isdABCDEFGH-encoded heme uptake system (e.g., IsdB). Taken together, IsrR establishes a link between the iron limitation response and the virulence in S. aureus.
Keywords: SaeRS TCS; Staphylococcus aureus; exoproteome; heme uptake; hemolysin; iron limitation; regulatory RNA; sRNA; secretome; virulence factor.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

