Phenotypic plasticity in cell elongation among closely related bacterial species
- PMID: 40456757
- PMCID: PMC12130487
- DOI: 10.1038/s41467-025-60005-y
Phenotypic plasticity in cell elongation among closely related bacterial species
Abstract
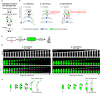
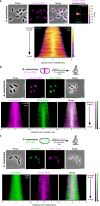
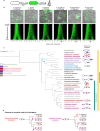
Cell elongation in bacteria has been studied over many decades, in part because its underlying mechanisms are targets of numerous antibiotics. While multiple elongation modes have been described, little is known about how these strategies vary across species and in response to evolutionary and environmental influences. Here, we use fluorescent D-amino acids to track the spatiotemporal dynamics of bacterial cell elongation, revealing unsuspected diversity of elongation modes among closely related species of the family Caulobacteraceae. We identify species-specific combinations of dispersed, midcell and polar elongation that can be either unidirectional or bidirectional. Using genetic, cell biology, and phylogenetic approaches, we demonstrate that evolution of unidirectional-midcell elongation is accompanied by changes in the localization of the peptidoglycan synthase PBP2. Our findings reveal high phenotypic plasticity in elongation mechanisms, with implications for our understanding of bacterial growth and evolution.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



Update of
-
Phenotypic plasticity in bacterial elongation among closely related species.bioRxiv [Preprint]. 2024 Nov 7:2024.11.07.622495. doi: 10.1101/2024.11.07.622495. bioRxiv. 2024. Update in: Nat Commun. 2025 Jun 2;16(1):5099. doi: 10.1038/s41467-025-60005-y. PMID: 39574579 Free PMC article. Updated. Preprint.
References
-
- Egan, A. J., Errington, J. & Vollmer, W. Regulation of peptidoglycan synthesis and remodelling. Nat. Rev. Microbiol.18, 446–460 (2020). - PubMed
MeSH terms
Substances
Grants and funding
- R35GM122556/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- R35 GM122556/GM/NIGMS NIH HHS/United States
- R35 GM136365/GM/NIGMS NIH HHS/United States
- R35GM136365/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- Junior 1 and Junior 2/Fonds de Recherche du Québec - Santé (Fonds de la recherche en sante du Quebec)
LinkOut - more resources
Full Text Sources

