NOTCH1 signaling is dysregulated by loss of the deubiquitinase USP28 with del(11q), uncovering USP28 inhibition as novel therapeutic target in CLL
- PMID: 40456839
- PMCID: PMC12310537
- DOI: 10.1038/s41375-025-02632-4
NOTCH1 signaling is dysregulated by loss of the deubiquitinase USP28 with del(11q), uncovering USP28 inhibition as novel therapeutic target in CLL
Abstract
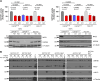
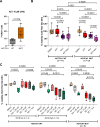
Aberrant active NOTCH1 signaling is a key pathogenic factor in chronic lymphocytic leukemia (CLL), detectable in half of patients and associated with disease progression. While some cases of active NOTCH1 signaling can be explained by mutations in NOTCH1 or its regulators, like FBXW7, alternative mechanisms remain elusive. Here, we identified the deubiquitinase USP28 as regulator of NOTCH1 signaling in CLL. Notably, USP28 is located within the frequently deleted chr11q23 region and is deleted in 90% of del(11q) patients, resulting in its decreased expression. USP28 interacts with the NOTCH1 intracellular domain (NICD) independently of FBXW7 and the NICD-PEST domain, stabilizing NICD and enhancing NOTCH1 signaling. Integrating RBPJ-occupied genes in HG3 cells, RNA-Seq of USP28WT/KO cells and gene expression from del(11q) CLL patients, we identified 15 NOTCH1 target genes specifically dysregulated by deletion of USP28 and del(11q) potentially influencing CLL pathogenesis. Pharmacological inhibition of USP28 with the small molecule AZ1 suppressed NOTCH1 activation in primary CLL cells. AZ1 combined with the BCL-2 inhibitor venetoclax reduced CLL cell viability, particularly in samples with high NOTCH1 activity. Our findings highlight USP28 as promising therapeutic target and provide a rationale for combined inhibition of USP28 and BCL-2 in CLL patients with active NOTCH1 signaling.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics approval: Collection and usage of primary cells from CLL patients has been approved by the Ethics Committee of Ulm University (Vote 242/20). All methods were performed in accordance with the relevant guidelines and regulations. Informed consent: All patients had given written informed consent to the collection and usage of their cells for research purposes according to the Declaration of Helsinki.
Figures
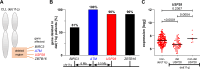


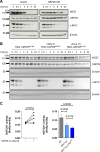
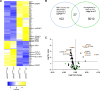
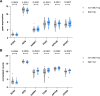
References
-
- Döhner H, Stilgenbauer S, Benner A, Leupolt E, Kröber A, Bullinger L, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–6. - PubMed
MeSH terms
Substances
Grants and funding
- 70114291/Deutsche Krebshilfe (German Cancer Aid)
- 70114289/Deutsche Krebshilfe (German Cancer Aid)
- SFB1074 projects B1 and B2/Deutsche Forschungsgemeinschaft (German Research Foundation)
- SFB1074 project A3/Deutsche Forschungsgemeinschaft (German Research Foundation)
- SFB1506 project A5/Deutsche Forschungsgemeinschaft (German Research Foundation)
LinkOut - more resources
Full Text Sources
Research Materials

