Neuronal aging causes mislocalization of splicing proteins and unchecked cellular stress
- PMID: 40456907
- PMCID: PMC12148940
- DOI: 10.1038/s41593-025-01952-z
Neuronal aging causes mislocalization of splicing proteins and unchecked cellular stress
Abstract
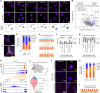
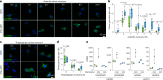
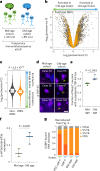
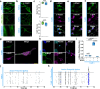
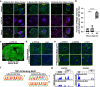
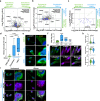
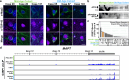
Aging is one of the most prominent risk factors for neurodegeneration, yet the molecular mechanisms underlying the deterioration of old neurons are mostly unknown. To efficiently study neurodegeneration in the context of aging, we transdifferentiated primary human fibroblasts from aged healthy donors directly into neurons, which retained their aging hallmarks, and we verified key findings in aged human and mouse brain tissue. Here we show that aged neurons are broadly depleted of RNA-binding proteins, especially spliceosome components. Intriguingly, splicing proteins-like the dementia- and ALS-associated protein TDP-43-mislocalize to the cytoplasm in aged neurons, which leads to widespread alternative splicing. Cytoplasmic spliceosome components are typically recruited to stress granules, but aged neurons suffer from chronic cellular stress that prevents this sequestration. We link chronic stress to the malfunctioning ubiquitylation machinery, poor HSP90α chaperone activity and the failure to respond to new stress events. Together, our data demonstrate that aging-linked deterioration of RNA biology is a key driver of poor resiliency in aged neurons.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: G.W.Y. is a member of the Scientific Advisory Board of Jumpcode Genomics and is a cofounder, member of the Board of Directors, Scientific Advisory Board member, equity holder, and paid consultant for Eclipse BioInnovations. G.W.Y.’s interests have been reviewed and approved by the University of California San Diego, in accordance with its conflict-of-interest policies. All other authors declare no competing interests.
Figures










References
MeSH terms
Substances
Grants and funding
- R35 GM148339/GM/NIGMS NIH HHS/United States
- DGE-2038238/National Science Foundation (NSF)
- P30-CA014195/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R01 NS069566/NS/NINDS NIH HHS/United States
- R01-HG004659/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- U01 CA267031/CA/NCI NIH HHS/United States
- P30-AG068635/U.S. Department of Health & Human Services | NIH | National Institute on Aging (U.S. National Institute on Aging)
- 23-PDF-639/Amyotrophic Lateral Sclerosis Association (ALS Association)
- P30 AG068635/AG/NIA NIH HHS/United States
- R01 HG004659/HG/NHGRI NIH HHS/United States
- U01-CA267031/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R35-GM148339/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- T32 CA067754/CA/NCI NIH HHS/United States
- R01 NS103172/NS/NINDS NIH HHS/United States
- R01-NS103172/U.S. Department of Health & Human Services | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- P30 CA014195/CA/NCI NIH HHS/United States
- IL-2023-C2-L4/Target ALS (Target ALS Foundation)
- T32-CA067754/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

