CleanSeqU algorithm for decontamination of catheterized urine 16S rRNA sequencing data
- PMID: 40457019
- PMCID: PMC12130503
- DOI: 10.1038/s41598-025-98875-3
CleanSeqU algorithm for decontamination of catheterized urine 16S rRNA sequencing data
Abstract
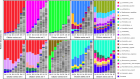
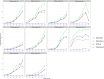
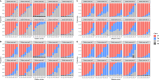
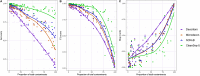
Contamination in low-biomass samples, such as urine, presents a major challenge for 16S rRNA gene sequencing, as extraneous DNA from reagents and the environment often obscures microbial signals. Existing in silico decontamination algorithms face limitations in accurately identifying and removing these contaminants. To address this issue, we developed CleanSeqU, a novel decontamination algorithm designed to enhance the accuracy of 16S rRNA gene sequencing data for catheterized urine samples. This approach is grounded in the principle that the compositional pattern of potential contaminant taxa remains similar between biological samples and blank controls. Also, the algorithm identifies potential contaminants based on ecological plausibility and custom blacklist. We evaluated CleanSeqU's performance using vaginal microbiome dilution experiments as a proxy for low-biomass urine samples and compared it to the Decontam, Microdecon, and SCRuB algorithm. CleanSeqU consistently outperformed Decontam, Microdecon, and SCRuB across various contamination levels, with superior accuracy, F1-scores, and reduced beta-dissimilarity. CleanSeqU improved specificity and positive predictive value by correctly identifying and removing a higher number of contaminant amplicon sequence variants (ASVs). Furthermore, the reduced alpha diversity in the decontaminated datasets suggests more precise contaminant elimination. With its practical use of a single blank extraction control per batch and adjustable decontamination rules, CleanSeqU provides an efficient and scalable solution that delivers accurate microbial profiles. Our findings highlight its potential to significantly advance urine microbiome research by delivering more accurate microbial profiles.
Keywords: 16S rRNA gene sequencing; Blank extraction control; Decontamination algorithms; Low biomass samples; Microbial contamination; Urine microbiome.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethics approval and consent to participants: All procedures involving the leftover of human vaginal samples and the study were approved by the Ethics Committee of GC Labs (GCL-2023-1075-02). All authors have provided their consent for publication.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

