KDM4C inhibition blocks tumor growth in basal breast cancer by promoting cathepsin L-mediated histone H3 cleavage
- PMID: 40457074
- PMCID: PMC12165855
- DOI: 10.1038/s41588-025-02197-z
KDM4C inhibition blocks tumor growth in basal breast cancer by promoting cathepsin L-mediated histone H3 cleavage
Abstract
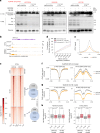
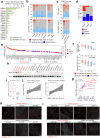
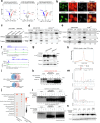
Basal breast cancer is a subtype with a poor prognosis in need of more effective therapeutic approaches. Here we describe a unique role for the KDM4C histone lysine demethylase in KDM4C-amplified basal breast cancers, where KDM4C inhibition reshapes chromatin and transcriptomic landscapes without substantial alterations of its canonical substrates, trimethylated histone H3 lysine 9 (H3K9me3) and lysine 36 (H3K36me3). Rather, KDM4C loss causes proteolytic cleavage of histone H3 mediated by cathepsin L (CTSL), resulting in decreased glutamate-cysteine ligase expression and increased reactive oxygen species. CTSL is recruited to the chromatin by the grainyhead-like 2 (GRHL2) transcription factor that is methylated at lysine 453 following KDM4C inhibition, triggering CTSL histone clipping activity. Deletion of CTSL rescued KDM4-loss-mediated tumor suppression. Our study reveals a function for KDM4C that connects cellular redox regulation and chromatin remodeling.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: K.P. serves on the Scientific Advisory Board of the Susan G. Komen Foundation, the V Foundation and Ideaya Biosciences; is an adviser to Curie.Bio; holds equity in Antares and stock options in Ideaya Biosciences; receives sponsored research funding from Novartis and received honoraria from AstraZeneca and sale of Scorpion Therapeutics stocks upon their acquisition by Eli Lilly in the past 12 months. F.M. is a cofounder of and has equity in Harbinger Health, has equity in Zephyr AI, and serves as a consultant for Harbinger Health, Zephyr AI and Red Cell Partners. F.M. declares that none of these relationships are directly or indirectly related to the content of this paper. L.E.S. and M.E. are current employees of AstraZeneca. M.L.-N. is a current employee of InSphero. D.J.M. is a current employee of Veralox Therapeutics. A.J. is a current employee of Pfizer. J.J. is a current employee of Odyssey Therapeutics. The other authors declare no competing interests.
Figures













References
MeSH terms
Substances
Grants and funding
- R35 CA197623/CA/NCI NIH HHS/United States
- P01 CA250959/CA/NCI NIH HHS/United States
- CA197623/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- W81XWH-14-1-0212/United States Department of Defense | United States Army | Army Medical Command | Congressionally Directed Medical Research Programs (CDMRP)
- CA250959/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
LinkOut - more resources
Full Text Sources
Medical

